Silvia Zuffi
Model-based Metric 3D Shape and Motion Reconstruction of Wild Bottlenose Dolphins in Drone-Shot Videos
Apr 22, 2025
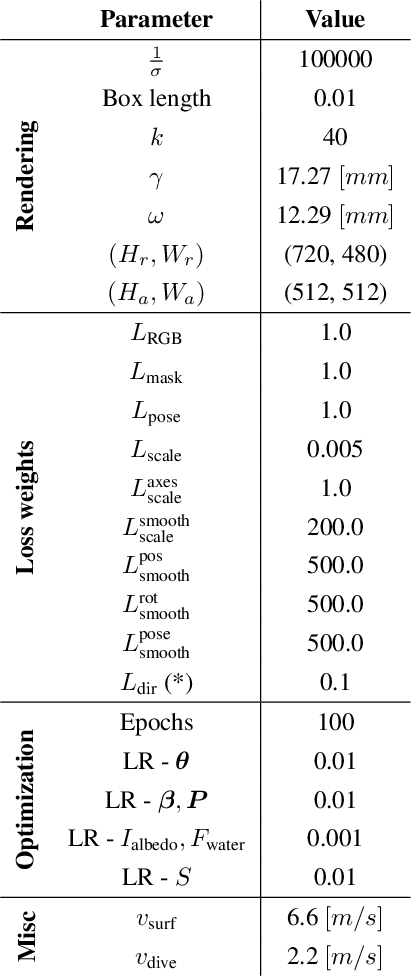
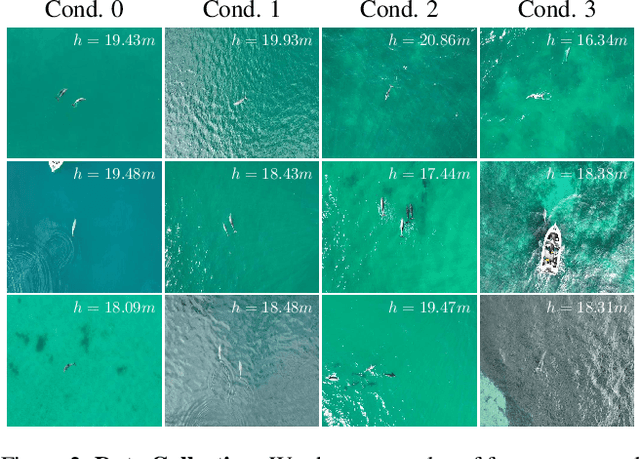
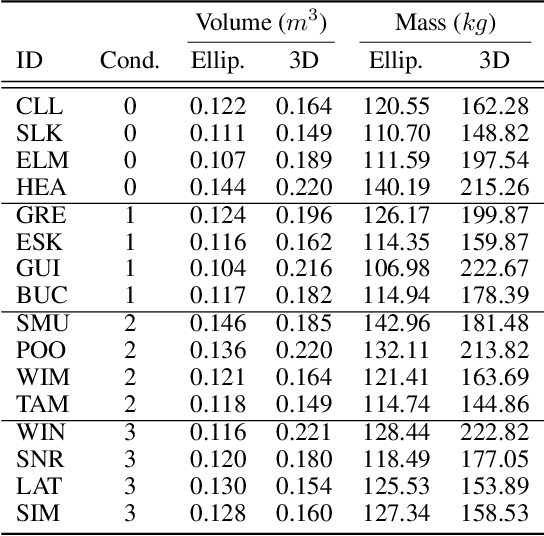
Abstract:We address the problem of estimating the metric 3D shape and motion of wild dolphins from monocular video, with the aim of assessing their body condition. While considerable progress has been made in reconstructing 3D models of terrestrial quadrupeds, aquatic animals remain unexplored due to the difficulty of observing them in their natural underwater environment. To address this, we propose a model-based approach that incorporates a transmission model to account for water-induced occlusion. We apply our method to video captured under different sea conditions. We estimate mass and volume, and compare our results to a manual 2D measurements-based method.
Generative Zoo
Dec 11, 2024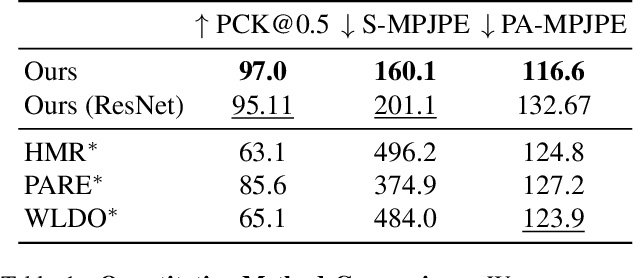
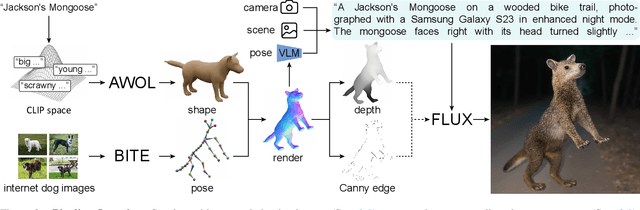
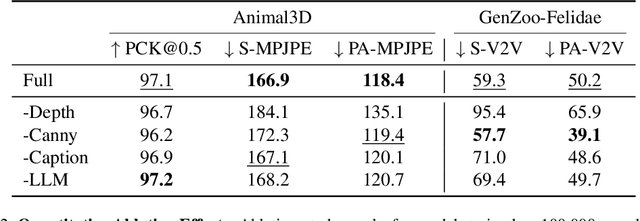

Abstract:The model-based estimation of 3D animal pose and shape from images enables computational modeling of animal behavior. Training models for this purpose requires large amounts of labeled image data with precise pose and shape annotations. However, capturing such data requires the use of multi-view or marker-based motion-capture systems, which are impractical to adapt to wild animals in situ and impossible to scale across a comprehensive set of animal species. Some have attempted to address the challenge of procuring training data by pseudo-labeling individual real-world images through manual 2D annotation, followed by 3D-parameter optimization to those labels. While this approach may produce silhouette-aligned samples, the obtained pose and shape parameters are often implausible due to the ill-posed nature of the monocular fitting problem. Sidestepping real-world ambiguity, others have designed complex synthetic-data-generation pipelines leveraging video-game engines and collections of artist-designed 3D assets. Such engines yield perfect ground-truth annotations but are often lacking in visual realism and require considerable manual effort to adapt to new species or environments. Motivated by these shortcomings, we propose an alternative approach to synthetic-data generation: rendering with a conditional image-generation model. We introduce a pipeline that samples a diverse set of poses and shapes for a variety of mammalian quadrupeds and generates realistic images with corresponding ground-truth pose and shape parameters. To demonstrate the scalability of our approach, we introduce GenZoo, a synthetic dataset containing one million images of distinct subjects. We train a 3D pose and shape regressor on GenZoo, which achieves state-of-the-art performance on a real-world animal pose and shape estimation benchmark, despite being trained solely on synthetic data. https://genzoo.is.tue.mpg.de
Reconstructing Animals and the Wild
Nov 27, 2024Abstract:The idea of 3D reconstruction as scene understanding is foundational in computer vision. Reconstructing 3D scenes from 2D visual observations requires strong priors to disambiguate structure. Much work has been focused on the anthropocentric, which, characterized by smooth surfaces, coherent normals, and regular edges, allows for the integration of strong geometric inductive biases. Here, we consider a more challenging problem where such assumptions do not hold: the reconstruction of natural scenes containing trees, bushes, boulders, and animals. While numerous works have attempted to tackle the problem of reconstructing animals in the wild, they have focused solely on the animal, neglecting environmental context. This limits their usefulness for analysis tasks, as animals exist inherently within the 3D world, and information is lost when environmental factors are disregarded. We propose a method to reconstruct natural scenes from single images. We base our approach on recent advances leveraging the strong world priors ingrained in Large Language Models and train an autoregressive model to decode a CLIP embedding into a structured compositional scene representation, encompassing both animals and the wild (RAW). To enable this, we propose a synthetic dataset comprising one million images and thousands of assets. Our approach, having been trained solely on synthetic data, generalizes to the task of reconstructing animals and their environments in real-world images. We will release our dataset and code to encourage future research at https://raw.is.tue.mpg.de/
Dessie: Disentanglement for Articulated 3D Horse Shape and Pose Estimation from Images
Oct 04, 2024Abstract:In recent years, 3D parametric animal models have been developed to aid in estimating 3D shape and pose from images and video. While progress has been made for humans, it's more challenging for animals due to limited annotated data. To address this, we introduce the first method using synthetic data generation and disentanglement to learn to regress 3D shape and pose. Focusing on horses, we use text-based texture generation and a synthetic data pipeline to create varied shapes, poses, and appearances, learning disentangled spaces. Our method, Dessie, surpasses existing 3D horse reconstruction methods and generalizes to other large animals like zebras, cows, and deer. See the project website at: \url{https://celiali.github.io/Dessie/}.
CLHOP: Combined Audio-Video Learning for Horse 3D Pose and Shape Estimation
Jul 01, 2024



Abstract:In the monocular setting, predicting 3D pose and shape of animals typically relies solely on visual information, which is highly under-constrained. In this work, we explore using audio to enhance 3D shape and motion recovery of horses from monocular video. We test our approach on two datasets: an indoor treadmill dataset for 3D evaluation and an outdoor dataset capturing diverse horse movements, the latter being a contribution to this study. Our results show that incorporating sound with visual data leads to more accurate and robust motion regression. This study is the first to investigate audio's role in 3D animal motion recovery.
AWOL: Analysis WithOut synthesis using Language
Apr 03, 2024Abstract:Many classical parametric 3D shape models exist, but creating novel shapes with such models requires expert knowledge of their parameters. For example, imagine creating a specific type of tree using procedural graphics or a new kind of animal from a statistical shape model. Our key idea is to leverage language to control such existing models to produce novel shapes. This involves learning a mapping between the latent space of a vision-language model and the parameter space of the 3D model, which we do using a small set of shape and text pairs. Our hypothesis is that mapping from language to parameters allows us to generate parameters for objects that were never seen during training. If the mapping between language and parameters is sufficiently smooth, then interpolation or generalization in language should translate appropriately into novel 3D shapes. We test our approach with two very different types of parametric shape models (quadrupeds and arboreal trees). We use a learned statistical shape model of quadrupeds and show that we can use text to generate new animals not present during training. In particular, we demonstrate state-of-the-art shape estimation of 3D dogs. This work also constitutes the first language-driven method for generating 3D trees. Finally, embedding images in the CLIP latent space enables us to generate animals and trees directly from images.
OSSO: Obtaining Skeletal Shape from Outside
Apr 21, 2022

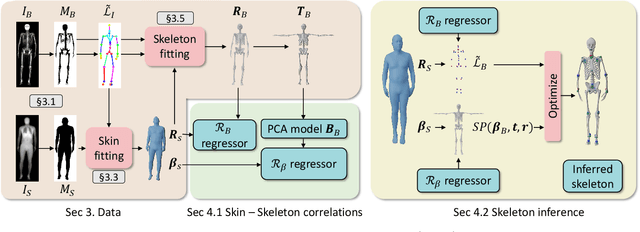
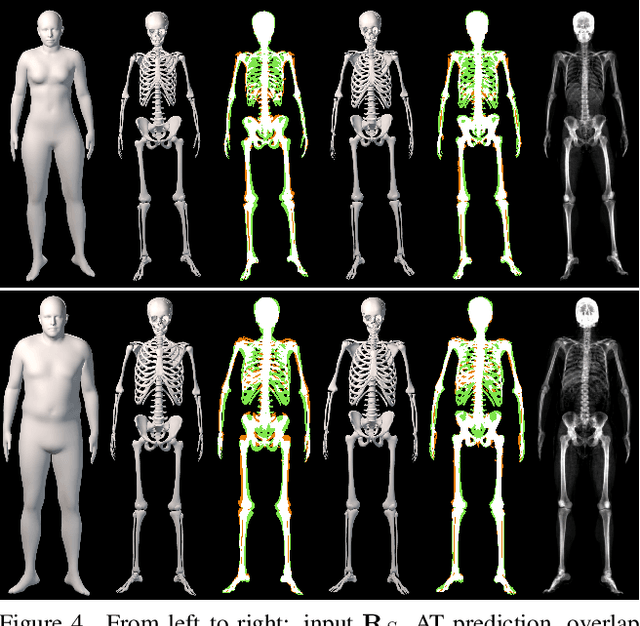
Abstract:We address the problem of inferring the anatomic skeleton of a person, in an arbitrary pose, from the 3D surface of the body; i.e. we predict the inside (bones) from the outside (skin). This has many applications in medicine and biomechanics. Existing state-of-the-art biomechanical skeletons are detailed but do not easily generalize to new subjects. Additionally, computer vision and graphics methods that predict skeletons are typically heuristic, not learned from data, do not leverage the full 3D body surface, and are not validated against ground truth. To our knowledge, our system, called OSSO (Obtaining Skeletal Shape from Outside), is the first to learn the mapping from the 3D body surface to the internal skeleton from real data. We do so using 1000 male and 1000 female dual-energy X-ray absorptiometry (DXA) scans. To these, we fit a parametric 3D body shape model (STAR) to capture the body surface and a novel part-based 3D skeleton model to capture the bones. This provides inside/outside training pairs. We model the statistical variation of full skeletons using PCA in a pose-normalized space. We then train a regressor from body shape parameters to skeleton shape parameters and refine the skeleton to satisfy constraints on physical plausibility. Given an arbitrary 3D body shape and pose, OSSO predicts a realistic skeleton inside. In contrast to previous work, we evaluate the accuracy of the skeleton shape quantitatively on held-out DXA scans, outperforming the state-of-the-art. We also show 3D skeleton prediction from varied and challenging 3D bodies. The code to infer a skeleton from a body shape is available for research at https://osso.is.tue.mpg.de/, and the dataset of paired outer surface (skin) and skeleton (bone) meshes is available as a Biobank Returned Dataset. This research has been conducted using the UK Biobank Resource.
BARC: Learning to Regress 3D Dog Shape from Images by Exploiting Breed Information
Mar 30, 2022
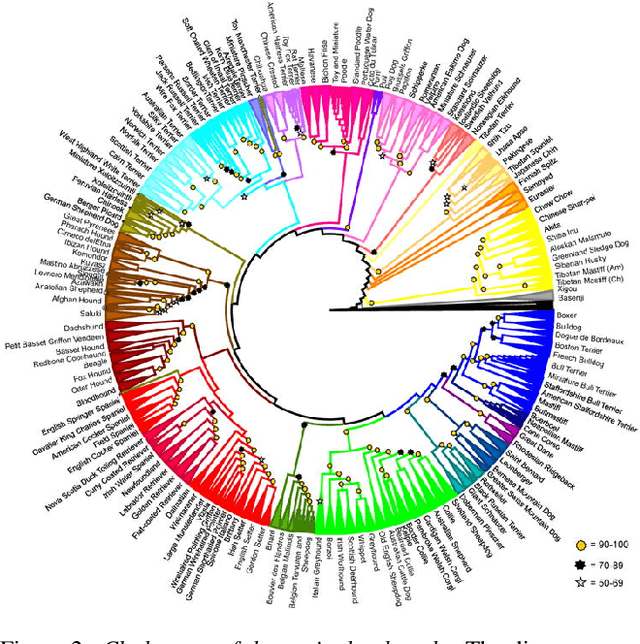

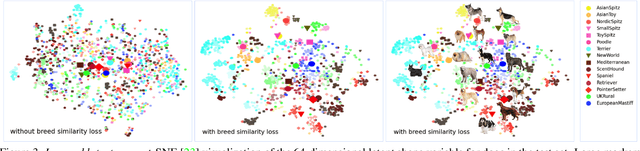
Abstract:Our goal is to recover the 3D shape and pose of dogs from a single image. This is a challenging task because dogs exhibit a wide range of shapes and appearances, and are highly articulated. Recent work has proposed to directly regress the SMAL animal model, with additional limb scale parameters, from images. Our method, called BARC (Breed-Augmented Regression using Classification), goes beyond prior work in several important ways. First, we modify the SMAL shape space to be more appropriate for representing dog shape. But, even with a better shape model, the problem of regressing dog shape from an image is still challenging because we lack paired images with 3D ground truth. To compensate for the lack of paired data, we formulate novel losses that exploit information about dog breeds. In particular, we exploit the fact that dogs of the same breed have similar body shapes. We formulate a novel breed similarity loss consisting of two parts: One term encourages the shape of dogs from the same breed to be more similar than dogs of different breeds. The second one, a breed classification loss, helps to produce recognizable breed-specific shapes. Through ablation studies, we find that our breed losses significantly improve shape accuracy over a baseline without them. We also compare BARC qualitatively to WLDO with a perceptual study and find that our approach produces dogs that are significantly more realistic. This work shows that a-priori information about genetic similarity can help to compensate for the lack of 3D training data. This concept may be applicable to other animal species or groups of species. Our code is publicly available for research purposes at https://barc.is.tue.mpg.de/.
Fish sounds: towards the evaluation of marine acoustic biodiversity through data-driven audio source separation
Jan 14, 2022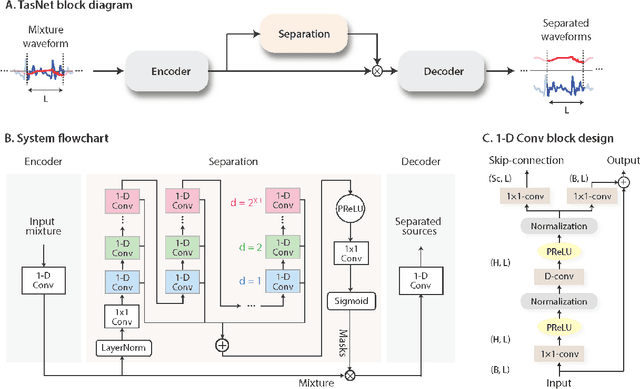
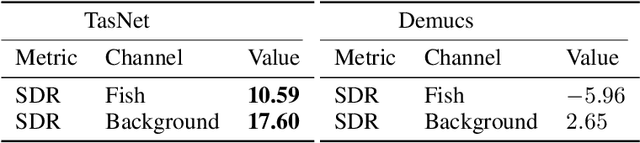
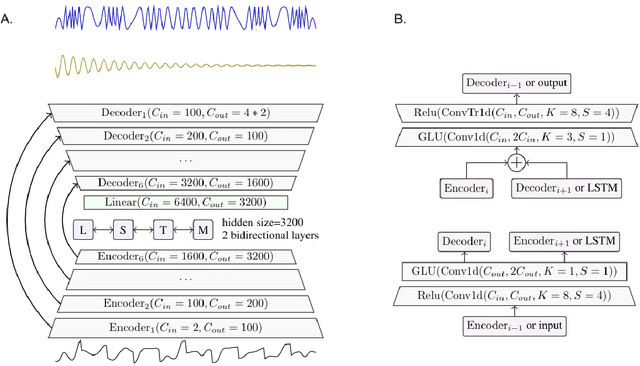
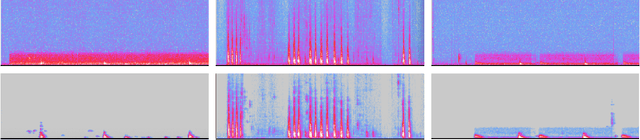
Abstract:The marine ecosystem is changing at an alarming rate, exhibiting biodiversity loss and the migration of tropical species to temperate basins. Monitoring the underwater environments and their inhabitants is of fundamental importance to understand the evolution of these systems and implement safeguard policies. However, assessing and tracking biodiversity is often a complex task, especially in large and uncontrolled environments, such as the oceans. One of the most popular and effective methods for monitoring marine biodiversity is passive acoustics monitoring (PAM), which employs hydrophones to capture underwater sound. Many aquatic animals produce sounds characteristic of their own species; these signals travel efficiently underwater and can be detected even at great distances. Furthermore, modern technologies are becoming more and more convenient and precise, allowing for very accurate and careful data acquisition. To date, audio captured with PAM devices is frequently manually processed by marine biologists and interpreted with traditional signal processing techniques for the detection of animal vocalizations. This is a challenging task, as PAM recordings are often over long periods of time. Moreover, one of the causes of biodiversity loss is sound pollution; in data obtained from regions with loud anthropic noise, it is hard to separate the artificial from the fish sound manually. Nowadays, machine learning and, in particular, deep learning represents the state of the art for processing audio signals. Specifically, sound separation networks are able to identify and separate human voices and musical instruments. In this work, we show that the same techniques can be successfully used to automatically extract fish vocalizations in PAM recordings, opening up the possibility for biodiversity monitoring at a large scale.
Seeing biodiversity: perspectives in machine learning for wildlife conservation
Oct 25, 2021
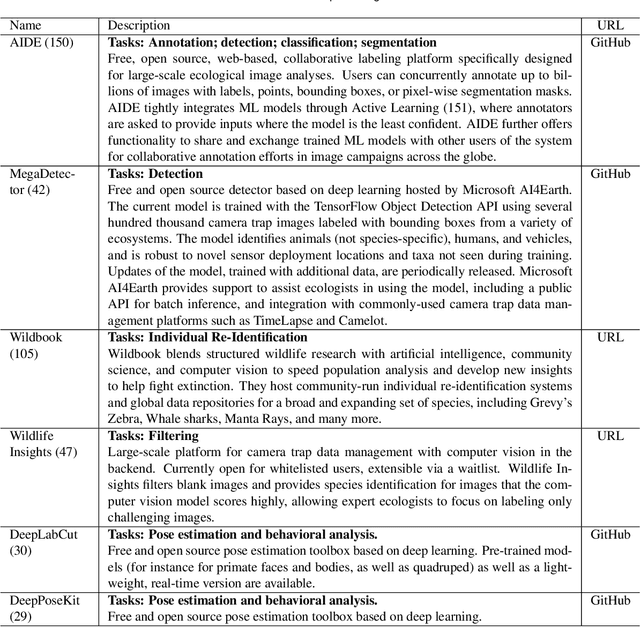
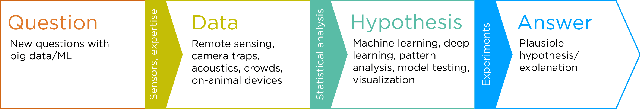
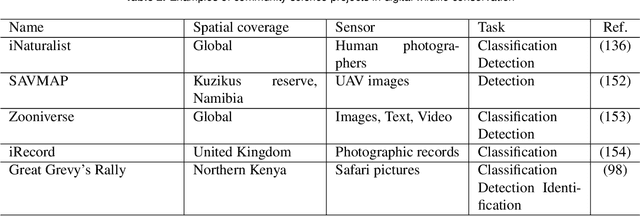
Abstract:Data acquisition in animal ecology is rapidly accelerating due to inexpensive and accessible sensors such as smartphones, drones, satellites, audio recorders and bio-logging devices. These new technologies and the data they generate hold great potential for large-scale environmental monitoring and understanding, but are limited by current data processing approaches which are inefficient in how they ingest, digest, and distill data into relevant information. We argue that machine learning, and especially deep learning approaches, can meet this analytic challenge to enhance our understanding, monitoring capacity, and conservation of wildlife species. Incorporating machine learning into ecological workflows could improve inputs for population and behavior models and eventually lead to integrated hybrid modeling tools, with ecological models acting as constraints for machine learning models and the latter providing data-supported insights. In essence, by combining new machine learning approaches with ecological domain knowledge, animal ecologists can capitalize on the abundance of data generated by modern sensor technologies in order to reliably estimate population abundances, study animal behavior and mitigate human/wildlife conflicts. To succeed, this approach will require close collaboration and cross-disciplinary education between the computer science and animal ecology communities in order to ensure the quality of machine learning approaches and train a new generation of data scientists in ecology and conservation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge