Benjamin Risse
ShelfOcc: Native 3D Supervision beyond LiDAR for Vision-Based Occupancy Estimation
Nov 19, 2025Abstract:Recent progress in self- and weakly supervised occupancy estimation has largely relied on 2D projection or rendering-based supervision, which suffers from geometric inconsistencies and severe depth bleeding. We thus introduce ShelfOcc, a vision-only method that overcomes these limitations without relying on LiDAR. ShelfOcc brings supervision into native 3D space by generating metrically consistent semantic voxel labels from video, enabling true 3D supervision without any additional sensors or manual 3D annotations. While recent vision-based 3D geometry foundation models provide a promising source of prior knowledge, they do not work out of the box as a prediction due to sparse or noisy and inconsistent geometry, especially in dynamic driving scenes. Our method introduces a dedicated framework that mitigates these issues by filtering and accumulating static geometry consistently across frames, handling dynamic content and propagating semantic information into a stable voxel representation. This data-centric shift in supervision for weakly/shelf-supervised occupancy estimation allows the use of essentially any SOTA occupancy model architecture without relying on LiDAR data. We argue that such high-quality supervision is essential for robust occupancy learning and constitutes an important complementary avenue to architectural innovation. On the Occ3D-nuScenes benchmark, ShelfOcc substantially outperforms all previous weakly/shelf-supervised methods (up to a 34% relative improvement), establishing a new data-driven direction for LiDAR-free 3D scene understanding.
GaussianFlowOcc: Sparse and Weakly Supervised Occupancy Estimation using Gaussian Splatting and Temporal Flow
Feb 25, 2025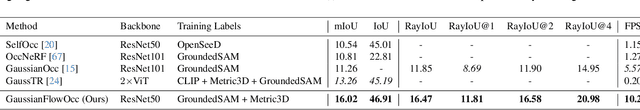
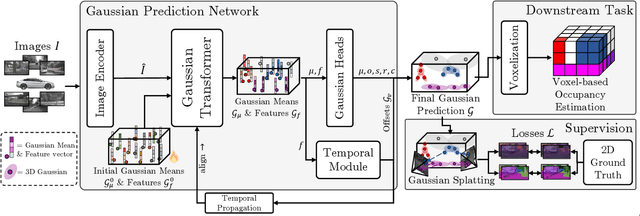
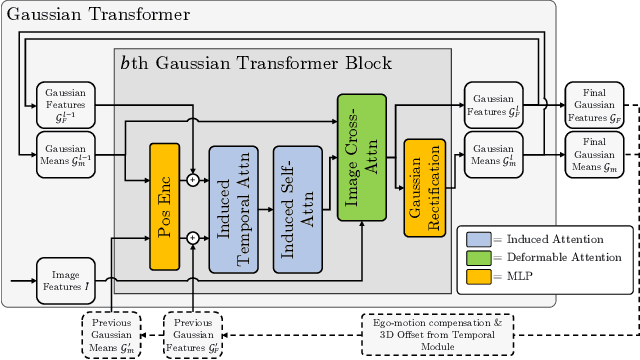
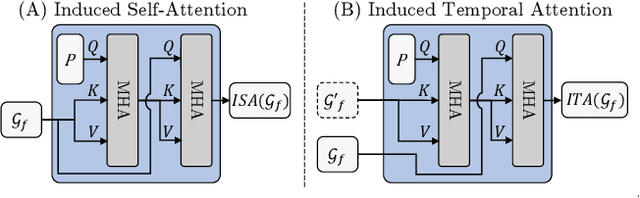
Abstract:Occupancy estimation has become a prominent task in 3D computer vision, particularly within the autonomous driving community. In this paper, we present a novel approach to occupancy estimation, termed GaussianFlowOcc, which is inspired by Gaussian Splatting and replaces traditional dense voxel grids with a sparse 3D Gaussian representation. Our efficient model architecture based on a Gaussian Transformer significantly reduces computational and memory requirements by eliminating the need for expensive 3D convolutions used with inefficient voxel-based representations that predominantly represent empty 3D spaces. GaussianFlowOcc effectively captures scene dynamics by estimating temporal flow for each Gaussian during the overall network training process, offering a straightforward solution to a complex problem that is often neglected by existing methods. Moreover, GaussianFlowOcc is designed for scalability, as it employs weak supervision and does not require costly dense 3D voxel annotations based on additional data (e.g., LiDAR). Through extensive experimentation, we demonstrate that GaussianFlowOcc significantly outperforms all previous methods for weakly supervised occupancy estimation on the nuScenes dataset while featuring an inference speed that is 50 times faster than current SOTA.
Learned Random Label Predictions as a Neural Network Complexity Metric
Nov 29, 2024Abstract:We empirically investigate the impact of learning randomly generated labels in parallel to class labels in supervised learning on memorization, model complexity, and generalization in deep neural networks. To this end, we introduce a multi-head network architecture as an extension of standard CNN architectures. Inspired by methods used in fair AI, our approach allows for the unlearning of random labels, preventing the network from memorizing individual samples. Based on the concept of Rademacher complexity, we first use our proposed method as a complexity metric to analyze the effects of common regularization techniques and challenge the traditional understanding of feature extraction and classification in CNNs. Second, we propose a novel regularizer that effectively reduces sample memorization. However, contrary to the predictions of classical statistical learning theory, we do not observe improvements in generalization.
Hatching-Box: Monitoring the Rearing Process of Drosophila Using an Embedded Imaging and in-vial Detection System
Nov 23, 2024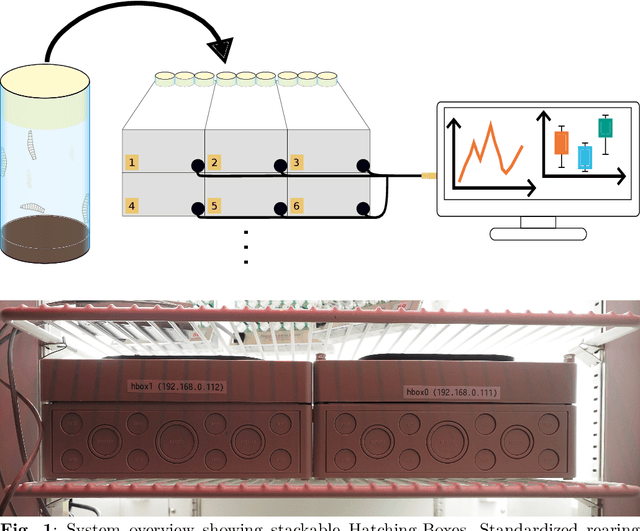
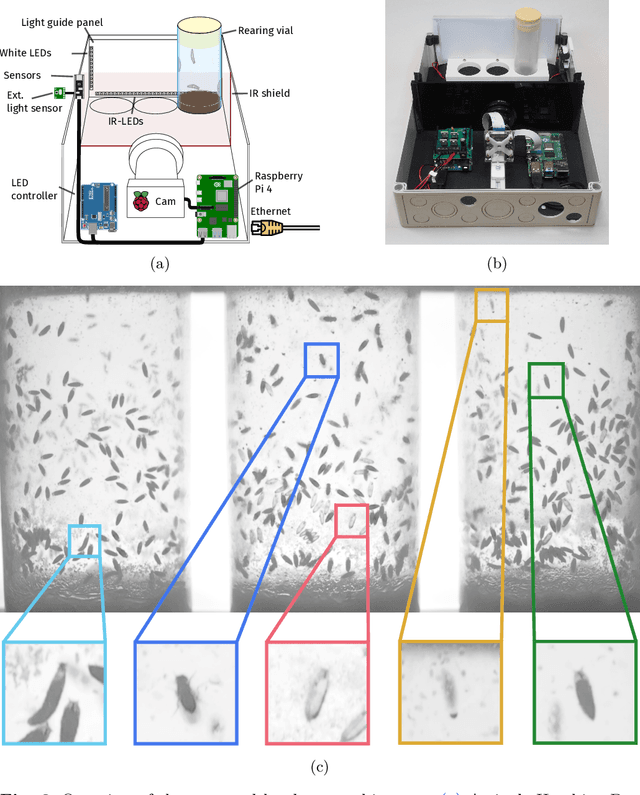
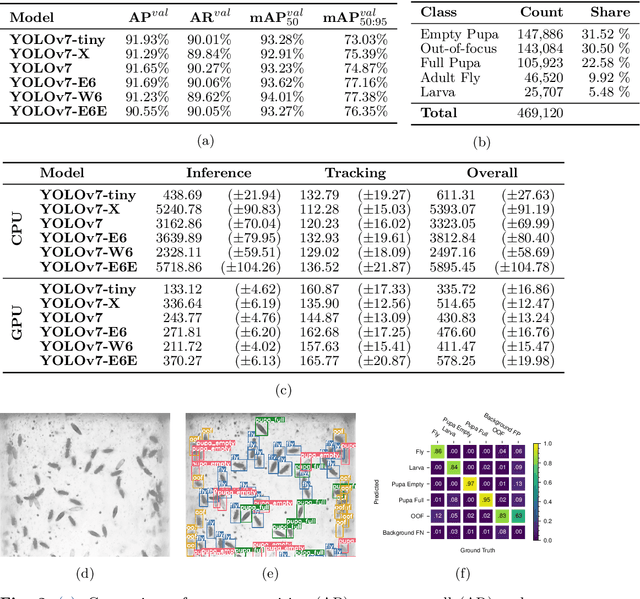
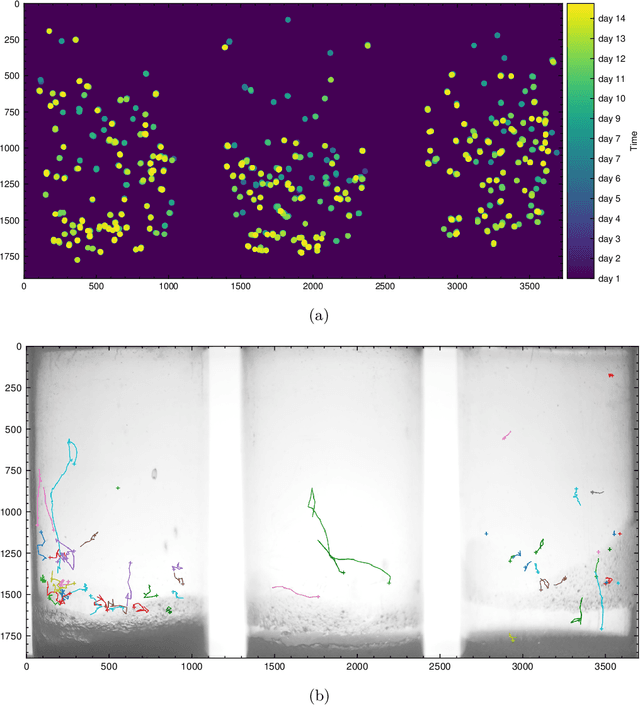
Abstract:In this paper we propose the Hatching-Box, a novel imaging and analysis system to automatically monitor and quantify the developmental behavior of Drosophila in standard rearing vials and during regular rearing routines, rendering explicit experiments obsolete. This is achieved by combining custom tailored imaging hardware with dedicated detection and tracking algorithms, enabling the quantification of larvae, filled/empty pupae and flies over multiple days. Given the affordable and reproducible design of the Hatching-Box in combination with our generic client/server-based software, the system can easily be scaled to monitor an arbitrary amount of rearing vials simultaneously. We evaluated our system on a curated image dataset comprising nearly 470,000 annotated objects and performed several studies on real world experiments. We successfully reproduced results from well-established circadian experiments by comparing the eclosion periods of wild type flies to the clock mutants $\textit{per}^{short}$, $\textit{per}^{long}$ and $\textit{per}^0$ without involvement of any manual labor. Furthermore we show, that the Hatching-Box is able to extract additional information about group behavior as well as to reconstruct the whole life-cycle of the individual specimens. These results not only demonstrate the applicability of our system for long-term experiments but also indicate its benefits for automated monitoring in the general cultivation process.
Towards Population Scale Testis Volume Segmentation in DIXON MRI
Oct 30, 2024Abstract:Testis size is known to be one of the main predictors of male fertility, usually assessed in clinical workup via palpation or imaging. Despite its potential, population-level evaluation of testicular volume using imaging remains underexplored. Previous studies, limited by small and biased datasets, have demonstrated the feasibility of machine learning for testis volume segmentation. This paper presents an evaluation of segmentation methods for testicular volume using Magnet Resonance Imaging data from the UKBiobank. The best model achieves a median dice score of $0.87$, compared to median dice score of $0.83$ for human interrater reliability on the same dataset, enabling large-scale annotation on a population scale for the first time. Our overall aim is to provide a trained model, comparative baseline methods, and annotated training data to enhance accessibility and reproducibility in testis MRI segmentation research.
deepmriprep: Voxel-based Morphometry (VBM) Preprocessing via Deep Neural Networks
Aug 20, 2024
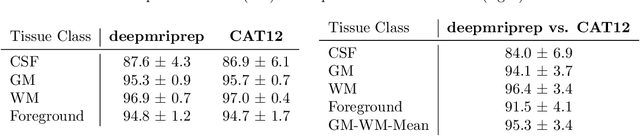


Abstract:Voxel-based Morphometry (VBM) has emerged as a powerful approach in neuroimaging research, utilized in over 7,000 studies since the year 2000. Using Magnetic Resonance Imaging (MRI) data, VBM assesses variations in the local density of brain tissue and examines its associations with biological and psychometric variables. Here, we present deepmriprep, a neural network-based pipeline that performs all necessary preprocessing steps for VBM analysis of T1-weighted MR images using deep neural networks. Utilizing the Graphics Processing Unit (GPU), deepmriprep is 37 times faster than CAT12, the leading VBM preprocessing toolbox. The proposed method matches CAT12 in accuracy for tissue segmentation and image registration across more than 100 datasets and shows strong correlations in VBM results. Tissue segmentation maps from deepmriprep have over 95% agreement with ground truth maps, and its non-linear registration, using supervised SYMNet, predicts smooth deformation fields comparable to CAT12. The high processing speed of deepmriprep enables rapid preprocessing of extensive datasets and thereby fosters the application of VBM analysis to large-scale neuroimaging studies and opens the door to real-time applications. Finally, deepmripreps straightforward, modular design enables researchers to easily understand, reuse, and advance the underlying methods, fostering further advancements in neuroimaging research. deepmriprep can be conveniently installed as a Python package and is publicly accessible at https://github.com/wwu-mmll/deepmriprep.
LangOcc: Self-Supervised Open Vocabulary Occupancy Estimation via Volume Rendering
Jul 25, 2024Abstract:The 3D occupancy estimation task has become an important challenge in the area of vision-based autonomous driving recently. However, most existing camera-based methods rely on costly 3D voxel labels or LiDAR scans for training, limiting their practicality and scalability. Moreover, most methods are tied to a predefined set of classes which they can detect. In this work we present a novel approach for open vocabulary occupancy estimation called LangOcc, that is trained only via camera images, and can detect arbitrary semantics via vision-language alignment. In particular, we distill the knowledge of the strong vision-language aligned encoder CLIP into a 3D occupancy model via differentiable volume rendering. Our model estimates vision-language aligned features in a 3D voxel grid using only images. It is trained in a self-supervised manner by rendering our estimations back to 2D space, where ground-truth features can be computed. This training mechanism automatically supervises the scene geometry, allowing for a straight-forward and powerful training method without any explicit geometry supervision. LangOcc outperforms LiDAR-supervised competitors in open vocabulary occupancy by a large margin, solely relying on vision-based training. We also achieve state-of-the-art results in self-supervised semantic occupancy estimation on the Occ3D-nuScenes dataset, despite not being limited to a specific set of categories, thus demonstrating the effectiveness of our proposed vision-language training.
OccFlowNet: Towards Self-supervised Occupancy Estimation via Differentiable Rendering and Occupancy Flow
Feb 20, 2024



Abstract:Semantic occupancy has recently gained significant traction as a prominent 3D scene representation. However, most existing methods rely on large and costly datasets with fine-grained 3D voxel labels for training, which limits their practicality and scalability, increasing the need for self-monitored learning in this domain. In this work, we present a novel approach to occupancy estimation inspired by neural radiance field (NeRF) using only 2D labels, which are considerably easier to acquire. In particular, we employ differentiable volumetric rendering to predict depth and semantic maps and train a 3D network based on 2D supervision only. To enhance geometric accuracy and increase the supervisory signal, we introduce temporal rendering of adjacent time steps. Additionally, we introduce occupancy flow as a mechanism to handle dynamic objects in the scene and ensure their temporal consistency. Through extensive experimentation we demonstrate that 2D supervision only is sufficient to achieve state-of-the-art performance compared to methods using 3D labels, while outperforming concurrent 2D approaches. When combining 2D supervision with 3D labels, temporal rendering and occupancy flow we outperform all previous occupancy estimation models significantly. We conclude that the proposed rendering supervision and occupancy flow advances occupancy estimation and further bridges the gap towards self-supervised learning in this domain.
pyAKI -- An Open Source Solution to Automated KDIGO classification
Jan 23, 2024Abstract:Acute Kidney Injury (AKI) is a frequent complication in critically ill patients, affecting up to 50% of patients in the intensive care units. The lack of standardized and open-source tools for applying the Kidney Disease Improving Global Outcomes (KDIGO) criteria to time series data has a negative impact on workload and study quality. This project introduces pyAKI, an open-source pipeline addressing this gap by providing a comprehensive solution for consistent KDIGO criteria implementation. The pyAKI pipeline was developed and validated using a subset of the Medical Information Mart for Intensive Care (MIMIC)-IV database, a commonly used database in critical care research. We defined a standardized data model in order to ensure reproducibility. Validation against expert annotations demonstrated pyAKI's robust performance in implementing KDIGO criteria. Comparative analysis revealed its ability to surpass the quality of human labels. This work introduces pyAKI as an open-source solution for implementing the KDIGO criteria for AKI diagnosis using time series data with high accuracy and performance.
Momentum-SAM: Sharpness Aware Minimization without Computational Overhead
Jan 22, 2024



Abstract:The recently proposed optimization algorithm for deep neural networks Sharpness Aware Minimization (SAM) suggests perturbing parameters before gradient calculation by a gradient ascent step to guide the optimization into parameter space regions of flat loss. While significant generalization improvements and thus reduction of overfitting could be demonstrated, the computational costs are doubled due to the additionally needed gradient calculation, making SAM unfeasible in case of limited computationally capacities. Motivated by Nesterov Accelerated Gradient (NAG) we propose Momentum-SAM (MSAM), which perturbs parameters in the direction of the accumulated momentum vector to achieve low sharpness without significant computational overhead or memory demands over SGD or Adam. We evaluate MSAM in detail and reveal insights on separable mechanisms of NAG, SAM and MSAM regarding training optimization and generalization. Code is available at https://github.com/MarlonBecker/MSAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge