Carlotta Barkhau
deepmriprep: Voxel-based Morphometry (VBM) Preprocessing via Deep Neural Networks
Aug 20, 2024
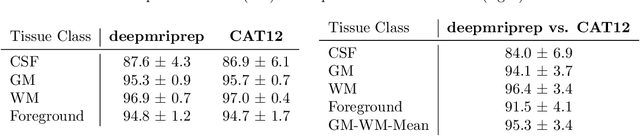


Abstract:Voxel-based Morphometry (VBM) has emerged as a powerful approach in neuroimaging research, utilized in over 7,000 studies since the year 2000. Using Magnetic Resonance Imaging (MRI) data, VBM assesses variations in the local density of brain tissue and examines its associations with biological and psychometric variables. Here, we present deepmriprep, a neural network-based pipeline that performs all necessary preprocessing steps for VBM analysis of T1-weighted MR images using deep neural networks. Utilizing the Graphics Processing Unit (GPU), deepmriprep is 37 times faster than CAT12, the leading VBM preprocessing toolbox. The proposed method matches CAT12 in accuracy for tissue segmentation and image registration across more than 100 datasets and shows strong correlations in VBM results. Tissue segmentation maps from deepmriprep have over 95% agreement with ground truth maps, and its non-linear registration, using supervised SYMNet, predicts smooth deformation fields comparable to CAT12. The high processing speed of deepmriprep enables rapid preprocessing of extensive datasets and thereby fosters the application of VBM analysis to large-scale neuroimaging studies and opens the door to real-time applications. Finally, deepmripreps straightforward, modular design enables researchers to easily understand, reuse, and advance the underlying methods, fostering further advancements in neuroimaging research. deepmriprep can be conveniently installed as a Python package and is publicly accessible at https://github.com/wwu-mmll/deepmriprep.
GateNet: A novel Neural Network Architecture for Automated Flow Cytometry Gating
Dec 12, 2023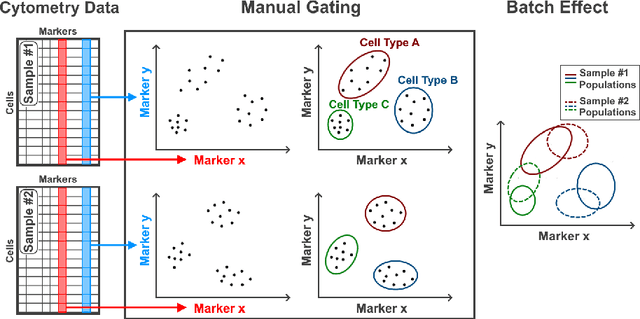
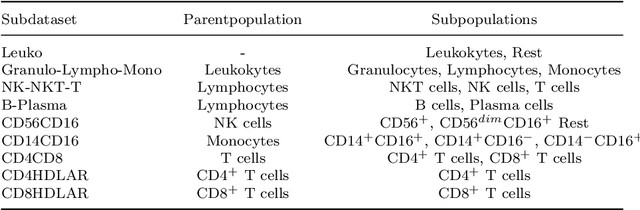
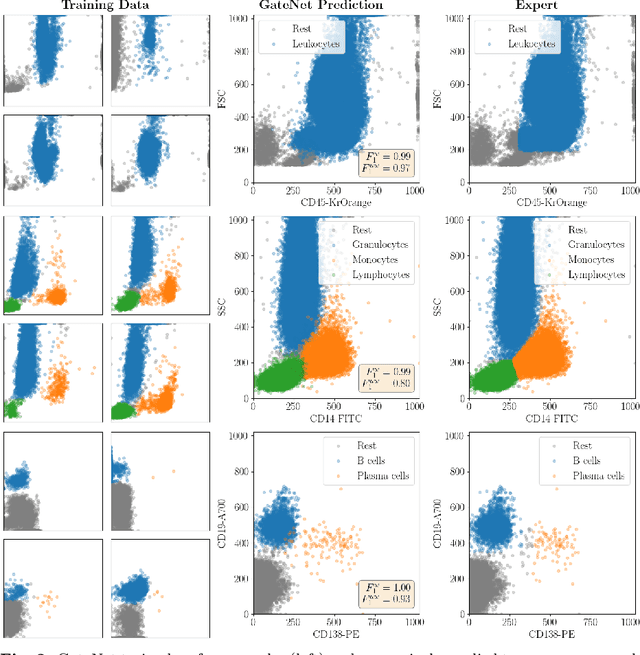
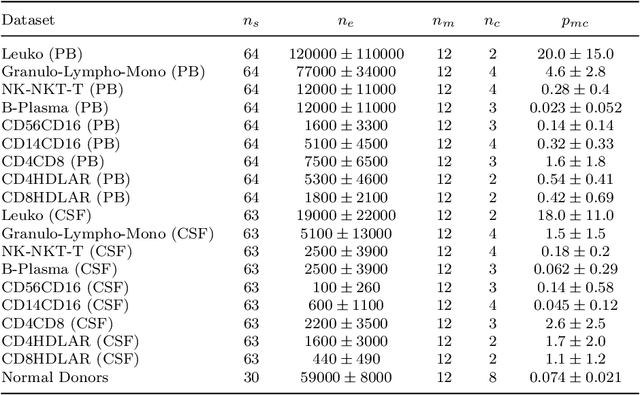
Abstract:Flow cytometry is widely used to identify cell populations in patient-derived fluids such as peripheral blood (PB) or cerebrospinal fluid (CSF). While ubiquitous in research and clinical practice, flow cytometry requires gating, i.e. cell type identification which requires labor-intensive and error-prone manual adjustments. To facilitate this process, we designed GateNet, the first neural network architecture enabling full end-to-end automated gating without the need to correct for batch effects. We train GateNet with over 8,000,000 events based on N=127 PB and CSF samples which were manually labeled independently by four experts. We show that for novel, unseen samples, GateNet achieves human-level performance (F1 score ranging from 0.910 to 0.997). In addition we apply GateNet to a publicly available dataset confirming generalization with an F1 score of 0.936. As our implementation utilizes graphics processing units (GPU), gating only needs 15 microseconds per event. Importantly, we also show that GateNet only requires ~10 samples to reach human-level performance, rendering it widely applicable in all domains of flow cytometry.
Deepbet: Fast brain extraction of T1-weighted MRI using Convolutional Neural Networks
Aug 14, 2023



Abstract:Brain extraction in magnetic resonance imaging (MRI) data is an important segmentation step in many neuroimaging preprocessing pipelines. Image segmentation is one of the research fields in which deep learning had the biggest impact in recent years enabling high precision segmentation with minimal compute. Consequently, traditional brain extraction methods are now being replaced by deep learning-based methods. Here, we used a unique dataset comprising 568 T1-weighted (T1w) MR images from 191 different studies in combination with cutting edge deep learning methods to build a fast, high-precision brain extraction tool called deepbet. deepbet uses LinkNet, a modern UNet architecture, in a two stage prediction process. This increases its segmentation performance, setting a novel state-of-the-art performance during cross-validation with a median Dice score (DSC) of 99.0% on unseen datasets, outperforming current state of the art models (DSC = 97.8% and DSC = 97.9%). While current methods are more sensitive to outliers, resulting in Dice scores as low as 76.5%, deepbet manages to achieve a Dice score of > 96.9% for all samples. Finally, our model accelerates brain extraction by a factor of ~10 compared to current methods, enabling the processing of one image in ~2 seconds on low level hardware.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge