Tilo Burghardt
Deep in the Jungle: Towards Automating Chimpanzee Population Estimation
Jan 30, 2026Abstract:The estimation of abundance and density in unmarked populations of great apes relies on statistical frameworks that require animal-to-camera distance measurements. In practice, acquiring these distances depends on labour-intensive manual interpretation of animal observations across large camera trap video corpora. This study introduces and evaluates an only sparsely explored alternative: the integration of computer vision-based monocular depth estimation (MDE) pipelines directly into ecological camera trap workflows for great ape conservation. Using a real-world dataset of 220 camera trap videos documenting a wild chimpanzee population, we combine two MDE models, Dense Prediction Transformers and Depth Anything, with multiple distance sampling strategies. These components are used to generate detection distance estimates, from which population density and abundance are inferred. Comparative analysis against manually derived ground-truth distances shows that calibrated DPT consistently outperforms Depth Anything. This advantage is observed in both distance estimation accuracy and downstream density and abundance inference. Nevertheless, both models exhibit systematic biases. We show that, given complex forest environments, they tend to overestimate detection distances and consequently underestimate density and abundance relative to conventional manual approaches. We further find that failures in animal detection across distance ranges are a primary factor limiting estimation accuracy. Overall, this work provides a case study that shows MDE-driven camera trap distance sampling is a viable and practical alternative to manual distance estimation. The proposed approach yields population estimates within 22% of those obtained using traditional methods.
Visual-textual Dermatoglyphic Animal Biometrics: A First Case Study on Panthera tigris
Dec 16, 2025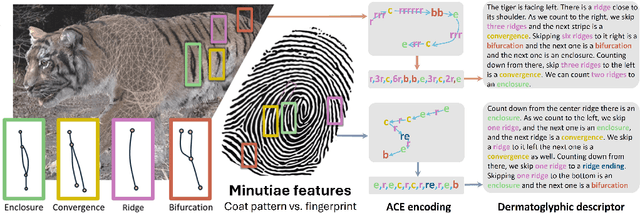
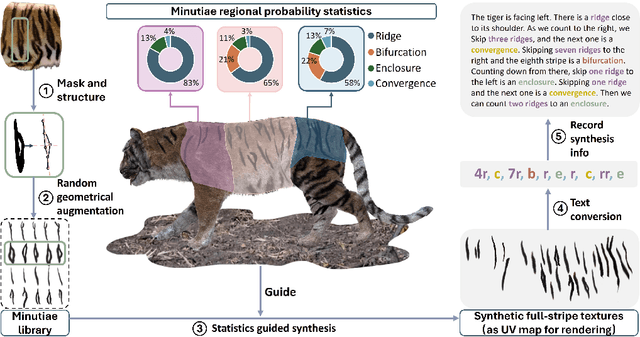
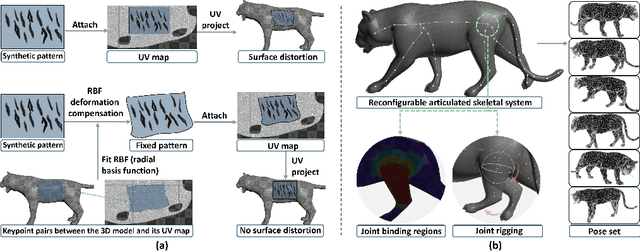
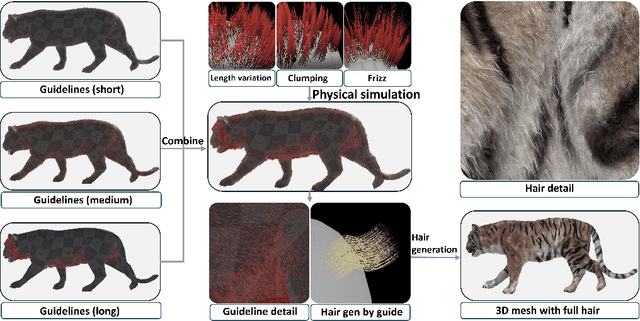
Abstract:Biologists have long combined visuals with textual field notes to re-identify (Re-ID) animals. Contemporary AI tools automate this for species with distinctive morphological features but remain largely image-based. Here, we extend Re-ID methodologies by incorporating precise dermatoglyphic textual descriptors-an approach used in forensics but new to ecology. We demonstrate that these specialist semantics abstract and encode animal coat topology using human-interpretable language tags. Drawing on 84,264 manually labelled minutiae across 3,355 images of 185 tigers (Panthera tigris), we evaluate this visual-textual methodology, revealing novel capabilities for cross-modal identity retrieval. To optimise performance, we developed a text-image co-synthesis pipeline to generate 'virtual individuals', each comprising dozens of life-like visuals paired with dermatoglyphic text. Benchmarking against real-world scenarios shows this augmentation significantly boosts AI accuracy in cross-modal retrieval while alleviating data scarcity. We conclude that dermatoglyphic language-guided biometrics can overcome vision-only limitations, enabling textual-to-visual identity recovery underpinned by human-verifiable matchings. This represents a significant advance towards explainability in Re-ID and a language-driven unification of descriptive modalities in ecological monitoring.
The SA-FARI Dataset: Segment Anything in Footage of Animals for Recognition and Identification
Nov 19, 2025Abstract:Automated video analysis is critical for wildlife conservation. A foundational task in this domain is multi-animal tracking (MAT), which underpins applications such as individual re-identification and behavior recognition. However, existing datasets are limited in scale, constrained to a few species, or lack sufficient temporal and geographical diversity - leaving no suitable benchmark for training general-purpose MAT models applicable across wild animal populations. To address this, we introduce SA-FARI, the largest open-source MAT dataset for wild animals. It comprises 11,609 camera trap videos collected over approximately 10 years (2014-2024) from 741 locations across 4 continents, spanning 99 species categories. Each video is exhaustively annotated culminating in ~46 hours of densely annotated footage containing 16,224 masklet identities and 942,702 individual bounding boxes, segmentation masks, and species labels. Alongside the task-specific annotations, we publish anonymized camera trap locations for each video. Finally, we present comprehensive benchmarks on SA-FARI using state-of-the-art vision-language models for detection and tracking, including SAM 3, evaluated with both species-specific and generic animal prompts. We also compare against vision-only methods developed specifically for wildlife analysis. SA-FARI is the first large-scale dataset to combine high species diversity, multi-region coverage, and high-quality spatio-temporal annotations, offering a new foundation for advancing generalizable multianimal tracking in the wild. The dataset is available at $\href{https://www.conservationxlabs.com/sa-fari}{\text{conservationxlabs.com/SA-FARI}}$.
Long-tailed Species Recognition in the NACTI Wildlife Dataset
Oct 24, 2025



Abstract:As most ''in the wild'' data collections of the natural world, the North America Camera Trap Images (NACTI) dataset shows severe long-tailed class imbalance, noting that the largest 'Head' class alone covers >50% of the 3.7M images in the corpus. Building on the PyTorch Wildlife model, we present a systematic study of Long-Tail Recognition methodologies for species recognition on the NACTI dataset covering experiments on various LTR loss functions plus LTR-sensitive regularisation. Our best configuration achieves 99.40% Top-1 accuracy on our NACTI test data split, substantially improving over a 95.51% baseline using standard cross-entropy with Adam. This also improves on previously reported top performance in MLWIC2 at 96.8% albeit using partly unpublished (potentially different) partitioning, optimiser, and evaluation protocols. To evaluate domain shifts (e.g. night-time captures, occlusion, motion-blur) towards other datasets we construct a Reduced-Bias Test set from the ENA-Detection dataset where our experimentally optimised long-tail enhanced model achieves leading 52.55% accuracy (up from 51.20% with WCE loss), demonstrating stronger generalisation capabilities under distribution shift. We document the consistent improvements of LTR-enhancing scheduler choices in this NACTI wildlife domain, particularly when in tandem with state-of-the-art LTR losses. We finally discuss qualitative and quantitative shortcomings that LTR methods cannot sufficiently address, including catastrophic breakdown for 'Tail' classes under severe domain shift. For maximum reproducibility we publish all dataset splits, key code, and full network weights.
Towards Application-Specific Evaluation of Vision Models: Case Studies in Ecology and Biology
May 05, 2025Abstract:Computer vision methods have demonstrated considerable potential to streamline ecological and biological workflows, with a growing number of datasets and models becoming available to the research community. However, these resources focus predominantly on evaluation using machine learning metrics, with relatively little emphasis on how their application impacts downstream analysis. We argue that models should be evaluated using application-specific metrics that directly represent model performance in the context of its final use case. To support this argument, we present two disparate case studies: (1) estimating chimpanzee abundance and density with camera trap distance sampling when using a video-based behaviour classifier and (2) estimating head rotation in pigeons using a 3D posture estimator. We show that even models with strong machine learning performance (e.g., 87% mAP) can yield data that leads to discrepancies in abundance estimates compared to expert-derived data. Similarly, the highest-performing models for posture estimation do not produce the most accurate inferences of gaze direction in pigeons. Motivated by these findings, we call for researchers to integrate application-specific metrics in ecological/biological datasets, allowing for models to be benchmarked in the context of their downstream application and to facilitate better integration of models into application workflows.
WildLive: Near Real-time Visual Wildlife Tracking onboard UAVs
Apr 15, 2025Abstract:Live tracking of wildlife via high-resolution video processing directly onboard drones is widely unexplored and most existing solutions rely on streaming video to ground stations to support navigation. Yet, both autonomous animal-reactive flight control beyond visual line of sight and/or mission-specific individual and behaviour recognition tasks rely to some degree on this capability. In response, we introduce WildLive -- a near real-time animal detection and tracking framework for high-resolution imagery running directly onboard uncrewed aerial vehicles (UAVs). The system performs multi-animal detection and tracking at 17fps+ for HD and 7fps+ on 4K video streams suitable for operation during higher altitude flights to minimise animal disturbance. Our system is optimised for Jetson Orin AGX onboard hardware. It integrates the efficiency of sparse optical flow tracking and mission-specific sampling with device-optimised and proven YOLO-driven object detection and segmentation techniques. Essentially, computational resource is focused onto spatio-temporal regions of high uncertainty to significantly improve UAV processing speeds without domain-specific loss of accuracy. Alongside, we introduce our WildLive dataset, which comprises 200k+ annotated animal instances across 19k+ frames from 4K UAV videos collected at the Ol Pejeta Conservancy in Kenya. All frames contain ground truth bounding boxes, segmentation masks, as well as individual tracklets and tracking point trajectories. We compare our system against current object tracking approaches including OC-SORT, ByteTrack, and SORT. Our materials are available at: https://dat-nguyenvn.github.io/WildLive/
MMLA: Multi-Environment, Multi-Species, Low-Altitude Aerial Footage Dataset
Apr 10, 2025Abstract:Real-time wildlife detection in drone imagery is critical for numerous applications, including animal ecology, conservation, and biodiversity monitoring. Low-altitude drone missions are effective for collecting fine-grained animal movement and behavior data, particularly if missions are automated for increased speed and consistency. However, little work exists on evaluating computer vision models on low-altitude aerial imagery and generalizability across different species and settings. To fill this gap, we present a novel multi-environment, multi-species, low-altitude aerial footage (MMLA) dataset. MMLA consists of drone footage collected across three diverse environments: Ol Pejeta Conservancy and Mpala Research Centre in Kenya, and The Wilds Conservation Center in Ohio, which includes five species: Plains zebras, Grevy's zebras, giraffes, onagers, and African Painted Dogs. We comprehensively evaluate three YOLO models (YOLOv5m, YOLOv8m, and YOLOv11m) for detecting animals. Results demonstrate significant performance disparities across locations and species-specific detection variations. Our work highlights the importance of evaluating detection algorithms across different environments for robust wildlife monitoring applications using drones.
Agglomerating Large Vision Encoders via Distillation for VFSS Segmentation
Apr 03, 2025Abstract:The deployment of foundation models for medical imaging has demonstrated considerable success. However, their training overheads associated with downstream tasks remain substantial due to the size of the image encoders employed, and the inference complexity is also significantly high. Although lightweight variants have been obtained for these foundation models, their performance is constrained by their limited model capacity and suboptimal training strategies. In order to achieve an improved tradeoff between complexity and performance, we propose a new framework to improve the performance of low complexity models via knowledge distillation from multiple large medical foundation models (e.g., MedSAM, RAD-DINO, MedCLIP), each specializing in different vision tasks, with the goal to effectively bridge the performance gap for medical image segmentation tasks. The agglomerated model demonstrates superior generalization across 12 segmentation tasks, whereas specialized models require explicit training for each task. Our approach achieved an average performance gain of 2\% in Dice coefficient compared to simple distillation.
The PanAf-FGBG Dataset: Understanding the Impact of Backgrounds in Wildlife Behaviour Recognition
Feb 28, 2025



Abstract:Computer vision analysis of camera trap video footage is essential for wildlife conservation, as captured behaviours offer some of the earliest indicators of changes in population health. Recently, several high-impact animal behaviour datasets and methods have been introduced to encourage their use; however, the role of behaviour-correlated background information and its significant effect on out-of-distribution generalisation remain unexplored. In response, we present the PanAf-FGBG dataset, featuring 20 hours of wild chimpanzee behaviours, recorded at over 350 individual camera locations. Uniquely, it pairs every video with a chimpanzee (referred to as a foreground video) with a corresponding background video (with no chimpanzee) from the same camera location. We present two views of the dataset: one with overlapping camera locations and one with disjoint locations. This setup enables, for the first time, direct evaluation of in-distribution and out-of-distribution conditions, and for the impact of backgrounds on behaviour recognition models to be quantified. All clips come with rich behavioural annotations and metadata including unique camera IDs and detailed textual scene descriptions. Additionally, we establish several baselines and present a highly effective latent-space normalisation technique that boosts out-of-distribution performance by +5.42% mAP for convolutional and +3.75% mAP for transformer-based models. Finally, we provide an in-depth analysis on the role of backgrounds in out-of-distribution behaviour recognition, including the so far unexplored impact of background durations (i.e., the count of background frames within foreground videos).
Tuning Vision Foundation Model via Test-Time Prompt-Guided Training for VFSS Segmentations
Jan 30, 2025



Abstract:Vision foundation models have demonstrated exceptional generalization capabilities in segmentation tasks for both generic and specialized images. However, a performance gap persists between foundation models and task-specific, specialized models. Fine-tuning foundation models on downstream datasets is often necessary to bridge this gap. Unfortunately, obtaining fully annotated ground truth for downstream datasets is both challenging and costly. To address this limitation, we propose a novel test-time training paradigm that enhances the performance of foundation models on downstream datasets without requiring full annotations. Specifically, our method employs simple point prompts to guide a test-time semi-self-supervised training task. The model learns by resolving the ambiguity of the point prompt through various augmentations. This approach directly tackles challenges in the medical imaging field, where acquiring annotations is both time-intensive and expensive. We conducted extensive experiments on our new Videofluoroscopy dataset (VFSS-5k) for the instance segmentation task, achieving an average Dice coefficient of 0.868 across 12 anatomies with a single model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge