Alberto M Gambaruto
Tuning Vision Foundation Model via Test-Time Prompt-Guided Training for VFSS Segmentations
Jan 30, 2025



Abstract:Vision foundation models have demonstrated exceptional generalization capabilities in segmentation tasks for both generic and specialized images. However, a performance gap persists between foundation models and task-specific, specialized models. Fine-tuning foundation models on downstream datasets is often necessary to bridge this gap. Unfortunately, obtaining fully annotated ground truth for downstream datasets is both challenging and costly. To address this limitation, we propose a novel test-time training paradigm that enhances the performance of foundation models on downstream datasets without requiring full annotations. Specifically, our method employs simple point prompts to guide a test-time semi-self-supervised training task. The model learns by resolving the ambiguity of the point prompt through various augmentations. This approach directly tackles challenges in the medical imaging field, where acquiring annotations is both time-intensive and expensive. We conducted extensive experiments on our new Videofluoroscopy dataset (VFSS-5k) for the instance segmentation task, achieving an average Dice coefficient of 0.868 across 12 anatomies with a single model.
RBF-PINN: Non-Fourier Positional Embedding in Physics-Informed Neural Networks
Feb 13, 2024Abstract:While many recent Physics-Informed Neural Networks (PINNs) variants have had considerable success in solving Partial Differential Equations, the empirical benefits of feature mapping drawn from the broader Neural Representations research have been largely overlooked. We highlight the limitations of widely used Fourier-based feature mapping in certain situations and suggest the use of the conditionally positive definite Radial Basis Function. The empirical findings demonstrate the effectiveness of our approach across a variety of forward and inverse problem cases. Our method can be seamlessly integrated into coordinate-based input neural networks and contribute to the wider field of PINNs research.
Training dynamics in Physics-Informed Neural Networks with feature mapping
Feb 10, 2024Abstract:Physics-Informed Neural Networks (PINNs) have emerged as an iconic machine learning approach for solving Partial Differential Equations (PDEs). Although its variants have achieved significant progress, the empirical success of utilising feature mapping from the wider Implicit Neural Representations studies has been substantially neglected. We investigate the training dynamics of PINNs with a feature mapping layer via the limiting Conjugate Kernel and Neural Tangent Kernel, which sheds light on the convergence and generalisation of the model. We also show the inadequacy of commonly used Fourier-based feature mapping in some scenarios and propose the conditional positive definite Radial Basis Function as a better alternative. The empirical results reveal the efficacy of our method in diverse forward and inverse problem sets. This simple technique can be easily implemented in coordinate input networks and benefits the broad PINNs research.
Video-SwinUNet: Spatio-temporal Deep Learning Framework for VFSS Instance Segmentation
Feb 22, 2023Abstract:This paper presents a deep learning framework for medical video segmentation. Convolution neural network (CNN) and transformer-based methods have achieved great milestones in medical image segmentation tasks due to their incredible semantic feature encoding and global information comprehension abilities. However, most existing approaches ignore a salient aspect of medical video data - the temporal dimension. Our proposed framework explicitly extracts features from neighbouring frames across the temporal dimension and incorporates them with a temporal feature blender, which then tokenises the high-level spatio-temporal feature to form a strong global feature encoded via a Swin Transformer. The final segmentation results are produced via a UNet-like encoder-decoder architecture. Our model outperforms other approaches by a significant margin and improves the segmentation benchmarks on the VFSS2022 dataset, achieving a dice coefficient of 0.8986 and 0.8186 for the two datasets tested. Our studies also show the efficacy of the temporal feature blending scheme and cross-dataset transferability of learned capabilities. Code and models are fully available at https://github.com/SimonZeng7108/Video-SwinUNet.
Video-TransUNet: Temporally Blended Vision Transformer for CT VFSS Instance Segmentation
Aug 22, 2022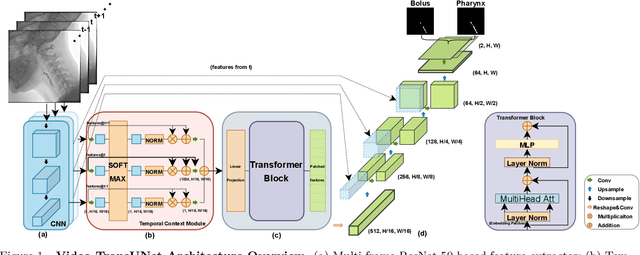
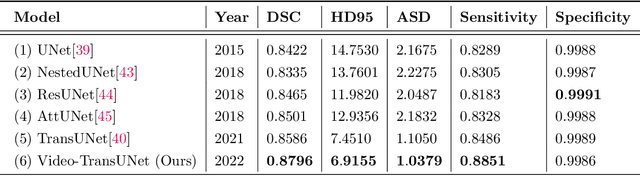
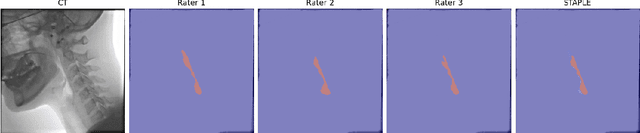

Abstract:We propose Video-TransUNet, a deep architecture for instance segmentation in medical CT videos constructed by integrating temporal feature blending into the TransUNet deep learning framework. In particular, our approach amalgamates strong frame representation via a ResNet CNN backbone, multi-frame feature blending via a Temporal Context Module (TCM), non-local attention via a Vision Transformer, and reconstructive capabilities for multiple targets via a UNet-based convolutional-deconvolutional architecture with multiple heads. We show that this new network design can significantly outperform other state-of-the-art systems when tested on the segmentation of bolus and pharynx/larynx in Videofluoroscopic Swallowing Study (VFSS) CT sequences. On our VFSS2022 dataset it achieves a dice coefficient of 0.8796 and an average surface distance of 1.0379 pixels. Note that tracking the pharyngeal bolus accurately is a particularly important application in clinical practice since it constitutes the primary method for diagnostics of swallowing impairment. Our findings suggest that the proposed model can indeed enhance the TransUNet architecture via exploiting temporal information and improving segmentation performance by a significant margin. We publish key source code, network weights, and ground truth annotations for simplified performance reproduction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge