Houda Chaabouni-Chouayakh
Teeth3DS: a benchmark for teeth segmentation and labeling from intra-oral 3D scans
Oct 12, 2022



Abstract:Teeth segmentation and labeling are critical components of Computer-Aided Dentistry (CAD) systems. Indeed, before any orthodontic or prosthetic treatment planning, a CAD system needs to first accurately segment and label each instance of teeth visible in the 3D dental scan, this is to avoid time-consuming manual adjustments by the dentist. Nevertheless, developing such an automated and accurate dental segmentation and labeling tool is very challenging, especially given the lack of publicly available datasets or benchmarks. This article introduces the first public benchmark, named Teeth3DS, which has been created in the frame of the 3DTeethSeg 2022 MICCAI challenge to boost the research field and inspire the 3D vision research community to work on intra-oral 3D scans analysis such as teeth identification, segmentation, labeling, 3D modeling and 3D reconstruction. Teeth3DS is made of 1800 intra-oral scans (23999 annotated teeth) collected from 900 patients covering the upper and lower jaws separately, acquired and validated by orthodontists/dental surgeons with more than 5 years of professional experience.
Self-Supervised Endoscopic Image Key-Points Matching
Aug 24, 2022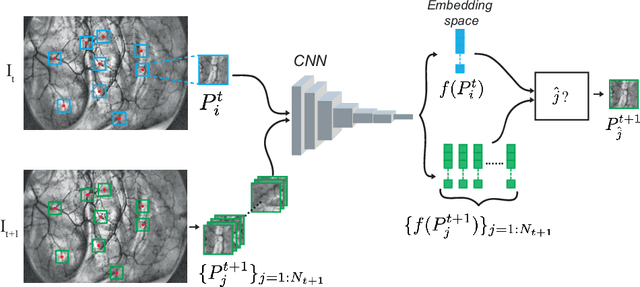
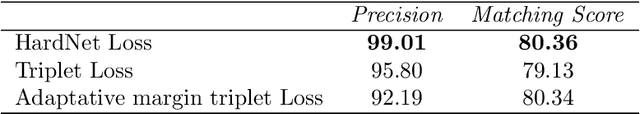
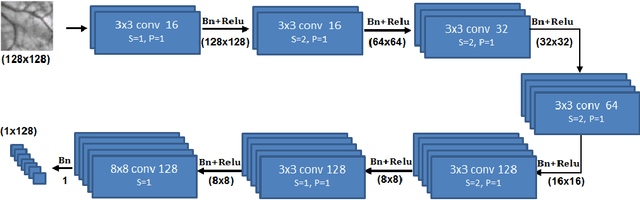
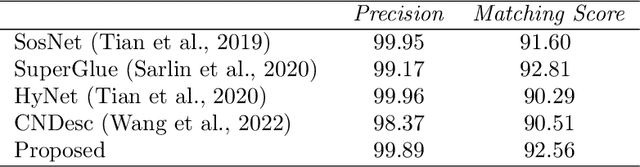
Abstract:Feature matching and finding correspondences between endoscopic images is a key step in many clinical applications such as patient follow-up and generation of panoramic image from clinical sequences for fast anomalies localization. Nonetheless, due to the high texture variability present in endoscopic images, the development of robust and accurate feature matching becomes a challenging task. Recently, deep learning techniques which deliver learned features extracted via convolutional neural networks (CNNs) have gained traction in a wide range of computer vision tasks. However, they all follow a supervised learning scheme where a large amount of annotated data is required to reach good performances, which is generally not always available for medical data databases. To overcome this limitation related to labeled data scarcity, the self-supervised learning paradigm has recently shown great success in a number of applications. This paper proposes a novel self-supervised approach for endoscopic image matching based on deep learning techniques. When compared to standard hand-crafted local feature descriptors, our method outperformed them in terms of precision and recall. Furthermore, our self-supervised descriptor provides a competitive performance in comparison to a selection of state-of-the-art deep learning based supervised methods in terms of precision and matching score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge