Chunlei Liu
Coordinate-Based Neural Representation Enabling Zero-Shot Learning for 3D Multiparametric Quantitative MRI
Oct 02, 2024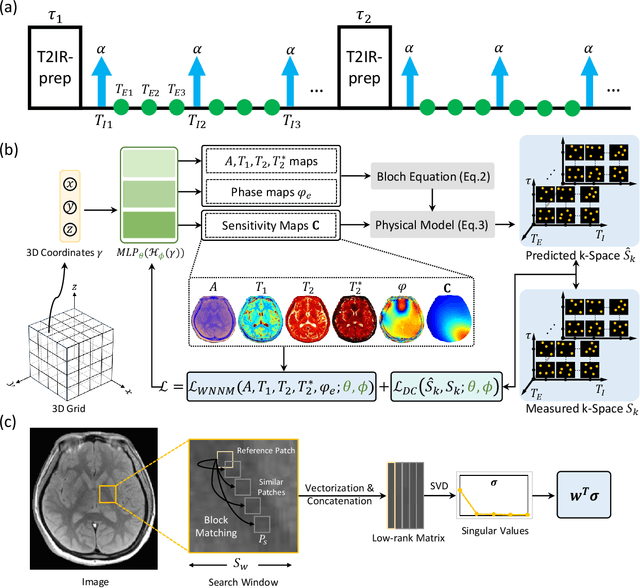

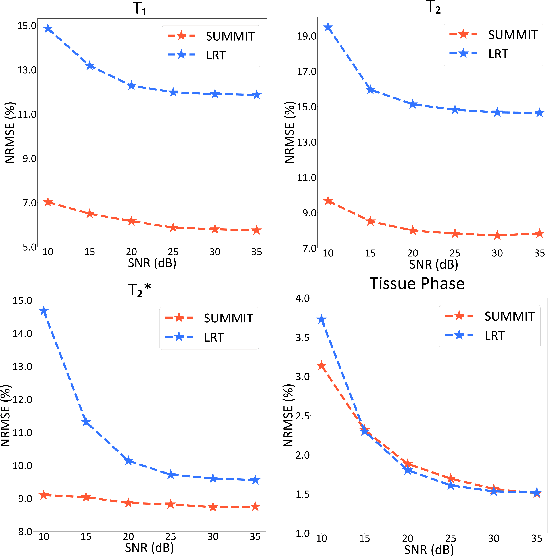
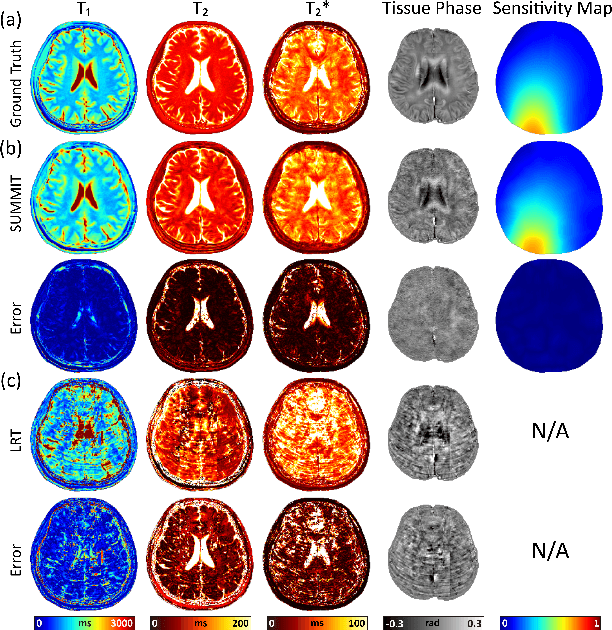
Abstract:Quantitative magnetic resonance imaging (qMRI) offers tissue-specific physical parameters with significant potential for neuroscience research and clinical practice. However, lengthy scan times for 3D multiparametric qMRI acquisition limit its clinical utility. Here, we propose SUMMIT, an innovative imaging methodology that includes data acquisition and an unsupervised reconstruction for simultaneous multiparametric qMRI. SUMMIT first encodes multiple important quantitative properties into highly undersampled k-space. It further leverages implicit neural representation incorporated with a dedicated physics model to reconstruct the desired multiparametric maps without needing external training datasets. SUMMIT delivers co-registered T1, T2, T2*, and quantitative susceptibility mapping. Extensive simulations and phantom imaging demonstrate SUMMIT's high accuracy. Additionally, the proposed unsupervised approach for qMRI reconstruction also introduces a novel zero-shot learning paradigm for multiparametric imaging applicable to various medical imaging modalities.
Interpretable deep learning in single-cell omics
Jan 11, 2024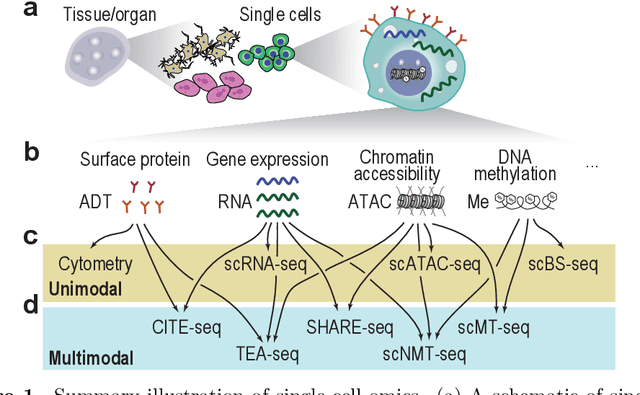
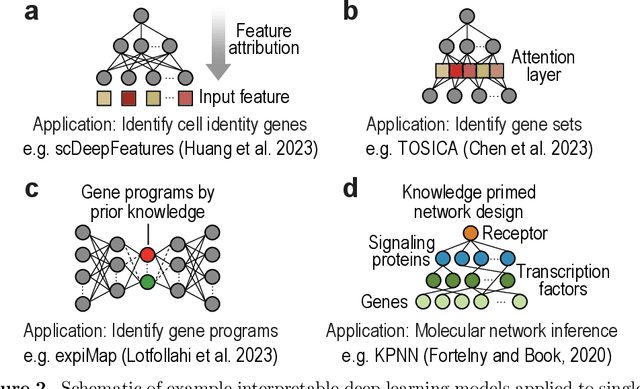
Abstract:Recent developments in single-cell omics technologies have enabled the quantification of molecular profiles in individual cells at an unparalleled resolution. Deep learning, a rapidly evolving sub-field of machine learning, has instilled a significant interest in single-cell omics research due to its remarkable success in analysing heterogeneous high-dimensional single-cell omics data. Nevertheless, the inherent multi-layer nonlinear architecture of deep learning models often makes them `black boxes' as the reasoning behind predictions is often unknown and not transparent to the user. This has stimulated an increasing body of research for addressing the lack of interpretability in deep learning models, especially in single-cell omics data analyses, where the identification and understanding of molecular regulators are crucial for interpreting model predictions and directing downstream experimental validations. In this work, we introduce the basics of single-cell omics technologies and the concept of interpretable deep learning. This is followed by a review of the recent interpretable deep learning models applied to various single-cell omics research. Lastly, we highlight the current limitations and discuss potential future directions. We anticipate this review to bring together the single-cell and machine learning research communities to foster future development and application of interpretable deep learning in single-cell omics research.
IMJENSE: Scan-specific Implicit Representation for Joint Coil Sensitivity and Image Estimation in Parallel MRI
Nov 21, 2023



Abstract:Parallel imaging is a commonly used technique to accelerate magnetic resonance imaging (MRI) data acquisition. Mathematically, parallel MRI reconstruction can be formulated as an inverse problem relating the sparsely sampled k-space measurements to the desired MRI image. Despite the success of many existing reconstruction algorithms, it remains a challenge to reliably reconstruct a high-quality image from highly reduced k-space measurements. Recently, implicit neural representation has emerged as a powerful paradigm to exploit the internal information and the physics of partially acquired data to generate the desired object. In this study, we introduced IMJENSE, a scan-specific implicit neural representation-based method for improving parallel MRI reconstruction. Specifically, the underlying MRI image and coil sensitivities were modeled as continuous functions of spatial coordinates, parameterized by neural networks and polynomials, respectively. The weights in the networks and coefficients in the polynomials were simultaneously learned directly from sparsely acquired k-space measurements, without fully sampled ground truth data for training. Benefiting from the powerful continuous representation and joint estimation of the MRI image and coil sensitivities, IMJENSE outperforms conventional image or k-space domain reconstruction algorithms. With extremely limited calibration data, IMJENSE is more stable than supervised calibrationless and calibration-based deep-learning methods. Results show that IMJENSE robustly reconstructs the images acquired at 5$\mathbf{\times}$ and 6$\mathbf{\times}$ accelerations with only 4 or 8 calibration lines in 2D Cartesian acquisitions, corresponding to 22.0% and 19.5% undersampling rates. The high-quality results and scanning specificity make the proposed method hold the potential for further accelerating the data acquisition of parallel MRI.
Towards Accurate Binary Neural Networks via Modeling Contextual Dependencies
Sep 03, 2022



Abstract:Existing Binary Neural Networks (BNNs) mainly operate on local convolutions with binarization function. However, such simple bit operations lack the ability of modeling contextual dependencies, which is critical for learning discriminative deep representations in vision models. In this work, we tackle this issue by presenting new designs of binary neural modules, which enables BNNs to learn effective contextual dependencies. First, we propose a binary multi-layer perceptron (MLP) block as an alternative to binary convolution blocks to directly model contextual dependencies. Both short-range and long-range feature dependencies are modeled by binary MLPs, where the former provides local inductive bias and the latter breaks limited receptive field in binary convolutions. Second, to improve the robustness of binary models with contextual dependencies, we compute the contextual dynamic embeddings to determine the binarization thresholds in general binary convolutional blocks. Armed with our binary MLP blocks and improved binary convolution, we build the BNNs with explicit Contextual Dependency modeling, termed as BCDNet. On the standard ImageNet-1K classification benchmark, the BCDNet achieves 72.3% Top-1 accuracy and outperforms leading binary methods by a large margin. In particular, the proposed BCDNet exceeds the state-of-the-art ReActNet-A by 2.9% Top-1 accuracy with similar operations. Our code is available at https://github.com/Sense-GVT/BCDN
Feature selection revisited in the single-cell era
Oct 27, 2021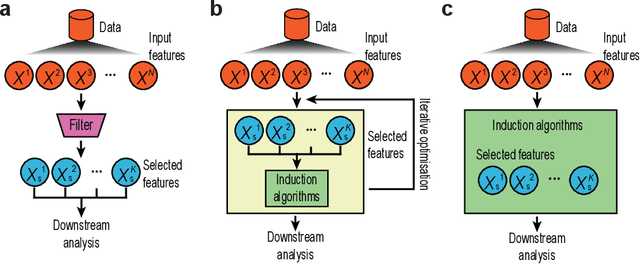
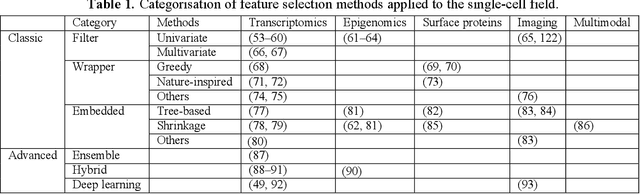
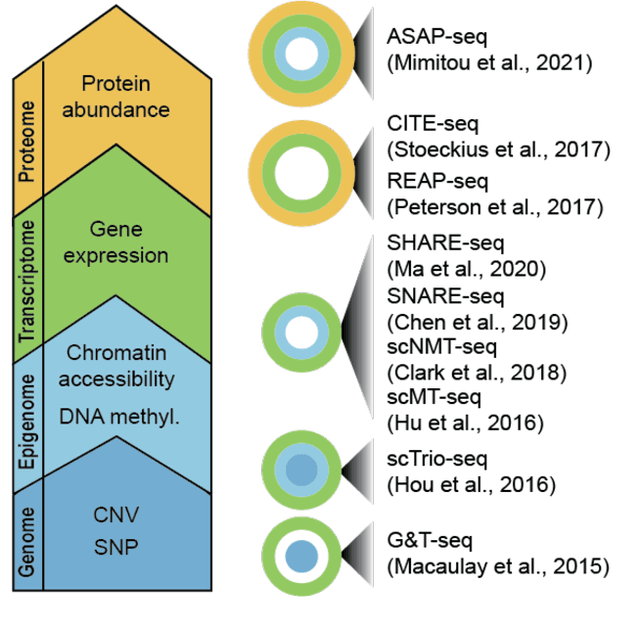
Abstract:Feature selection techniques are essential for high-dimensional data analysis. In the last two decades, their popularity has been fuelled by the increasing availability of high-throughput biomolecular data where high-dimensionality is a common data property. Recent advances in biotechnologies enable global profiling of various molecular and cellular features at single-cell resolution, resulting in large-scale datasets with increased complexity. These technological developments have led to a resurgence in feature selection research and application in the single-cell field. Here, we revisit feature selection techniques and summarise recent developments. We review their versatile application to a range of single-cell data types including those generated from traditional cytometry and imaging technologies and the latest array of single-cell omics technologies. We highlight some of the challenges and future directions on which feature selection could have a significant impact. Finally, we consider the scalability and make general recommendations on the utility of each type of feature selection method. We hope this review serves as a reference point to stimulate future research and application of feature selection in the single-cell era.
MoG-QSM: Model-based Generative Adversarial Deep Learning Network for Quantitative Susceptibility Mapping
Jan 21, 2021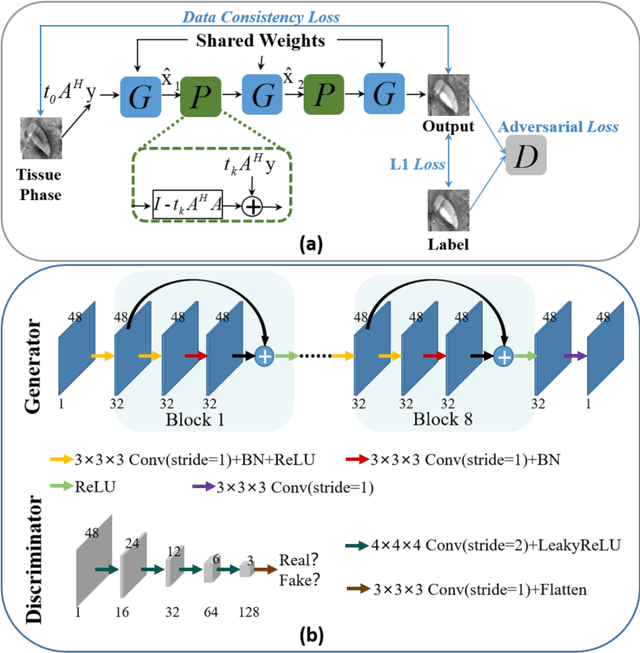
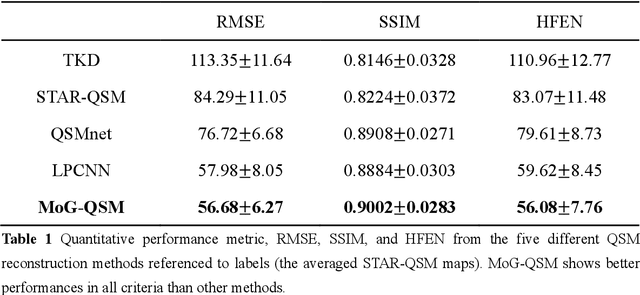
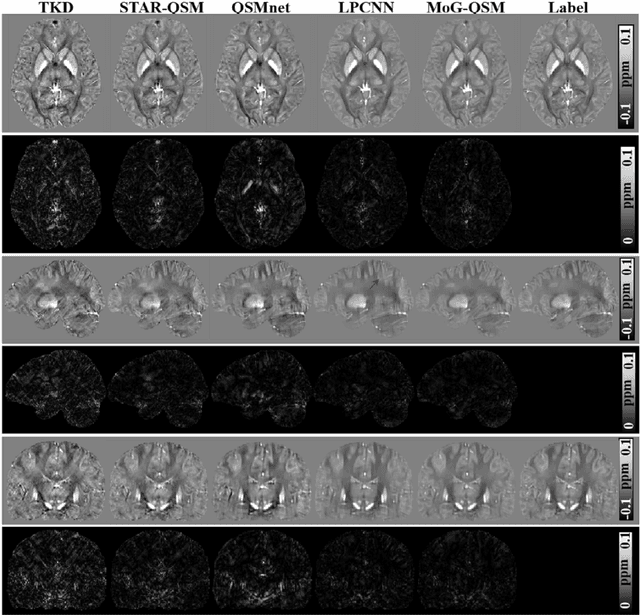
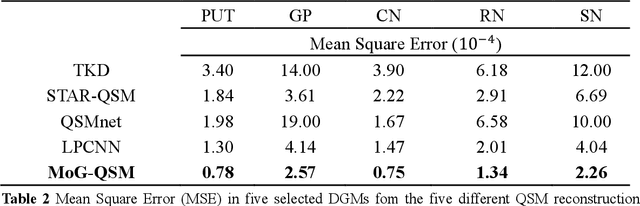
Abstract:Quantitative susceptibility mapping (QSM) estimates the underlying tissue magnetic susceptibility from the MRI gradient-echo phase signal and has demonstrated great potential in quantifying tissue susceptibility in various brain diseases. However, the intrinsic ill-posed inverse problem relating the tissue phase to the underlying susceptibility distribution affects the accuracy for quantifying tissue susceptibility. The resulting susceptibility map is known to suffer from noise amplification and streaking artifacts. To address these challenges, we propose a model-based framework that permeates benefits from generative adversarial networks to train a regularization term that contains prior information to constrain the solution of the inverse problem, referred to as MoG-QSM. A residual network leveraging a mixture of least-squares (LS) GAN and the L1 cost was trained as the generator to learn the prior information in susceptibility maps. A multilayer convolutional neural network was jointly trained to discriminate the quality of output images. MoG-QSM generates highly accurate susceptibility maps from single orientation phase maps. Quantitative evaluation parameters were compared with recently developed deep learning QSM methods and the results showed MoG-QSM achieves the best performance. Furthermore, a higher intraclass correlation coefficient (ICC) was obtained from MoG-QSM maps of the traveling subjects, demonstrating its potential for future applications, such as large cohorts of multi-center studies. MoG-QSM is also helpful for reliable longitudinal measurement of susceptibility time courses, enabling more precise monitoring for metal ion accumulation in neurodegenerative disorders.
GBCNs: Genetic Binary Convolutional Networks for Enhancing the Performance of 1-bit DCNNs
Nov 25, 2019



Abstract:Training 1-bit deep convolutional neural networks (DCNNs) is one of the most challenging problems in computer vision, because it is much easier to get trapped into local minima than conventional DCNNs. The reason lies in that the binarized kernels and activations of 1-bit DCNNs cause a significant accuracy loss and training inefficiency. To address this problem, we propose Genetic Binary Convolutional Networks (GBCNs) to optimize 1-bit DCNNs, by introducing a new balanced Genetic Algorithm (BGA) to improve the representational ability in an end-to-end framework. The BGA method is proposed to modify the binary process of GBCNs to alleviate the local minima problem, which can significantly improve the performance of 1-bit DCNNs. We develop a new BGA module that is generic and flexible, and can be easily incorporated into existing DCNNs, such asWideResNets and ResNets. Extensive experiments on the object classification tasks (CIFAR, ImageNet) validate the effectiveness of the proposed method. To highlight, our method shows strong generalization on the object recognition task, i.e., face recognition, facial and person re-identification.
Aggregation Signature for Small Object Tracking
Oct 24, 2019



Abstract:Small object tracking becomes an increasingly important task, which however has been largely unexplored in computer vision. The great challenges stem from the facts that: 1) small objects show extreme vague and variable appearances, and 2) they tend to be lost easier as compared to normal-sized ones due to the shaking of lens. In this paper, we propose a novel aggregation signature suitable for small object tracking, especially aiming for the challenge of sudden and large drift. We make three-fold contributions in this work. First, technically, we propose a new descriptor, named aggregation signature, based on saliency, able to represent highly distinctive features for small objects. Second, theoretically, we prove that the proposed signature matches the foreground object more accurately with a high probability. Third, experimentally, the aggregation signature achieves a high performance on multiple datasets, outperforming the state-of-the-art methods by large margins. Moreover, we contribute with two newly collected benchmark datasets, i.e., small90 and small112, for visually small object tracking. The datasets will be available in https://github.com/bczhangbczhang/.
Circulant Binary Convolutional Networks: Enhancing the Performance of 1-bit DCNNs with Circulant Back Propagation
Oct 24, 2019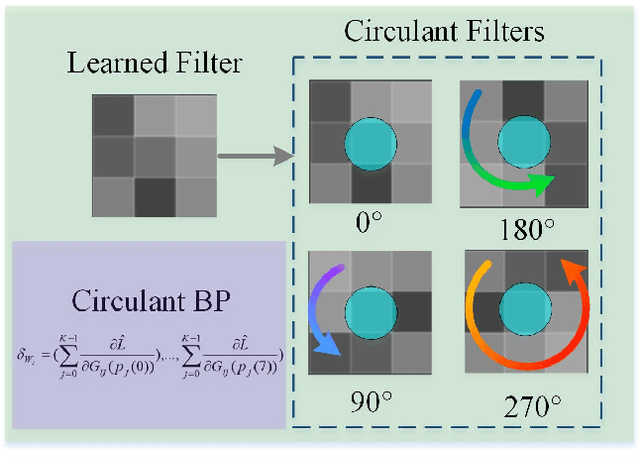

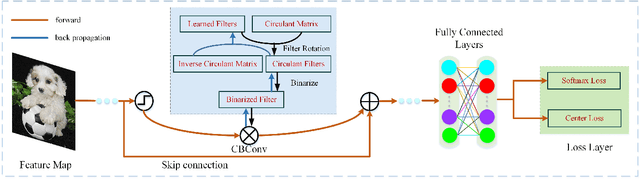
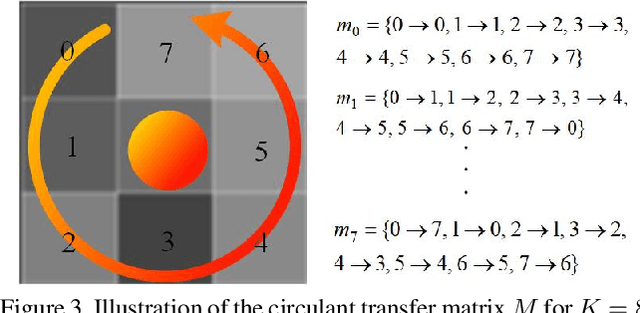
Abstract:The rapidly decreasing computation and memory cost has recently driven the success of many applications in the field of deep learning. Practical applications of deep learning in resource-limited hardware, such as embedded devices and smart phones, however, remain challenging. For binary convolutional networks, the reason lies in the degraded representation caused by binarizing full-precision filters. To address this problem, we propose new circulant filters (CiFs) and a circulant binary convolution (CBConv) to enhance the capacity of binarized convolutional features via our circulant back propagation (CBP). The CiFs can be easily incorporated into existing deep convolutional neural networks (DCNNs), which leads to new Circulant Binary Convolutional Networks (CBCNs). Extensive experiments confirm that the performance gap between the 1-bit and full-precision DCNNs is minimized by increasing the filter diversity, which further increases the representational ability in our networks. Our experiments on ImageNet show that CBCNs achieve 61.4% top-1 accuracy with ResNet18. Compared to the state-of-the-art such as XNOR, CBCNs can achieve up to 10% higher top-1 accuracy with more powerful representational ability.
* Published in CVPR2019
RBCN: Rectified Binary Convolutional Networks for Enhancing the Performance of 1-bit DCNNs
Sep 06, 2019



Abstract:Binarized convolutional neural networks (BCNNs) are widely used to improve memory and computation efficiency of deep convolutional neural networks (DCNNs) for mobile and AI chips based applications. However, current BCNNs are not able to fully explore their corresponding full-precision models, causing a significant performance gap between them. In this paper, we propose rectified binary convolutional networks (RBCNs), towards optimized BCNNs, by combining full-precision kernels and feature maps to rectify the binarization process in a unified framework. In particular, we use a GAN to train the 1-bit binary network with the guidance of its corresponding full-precision model, which significantly improves the performance of BCNNs. The rectified convolutional layers are generic and flexible, and can be easily incorporated into existing DCNNs such as WideResNets and ResNets. Extensive experiments demonstrate the superior performance of the proposed RBCNs over state-of-the-art BCNNs. In particular, our method shows strong generalization on the object tracking task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge