Pengyi Yang
Interpretable deep learning in single-cell omics
Jan 11, 2024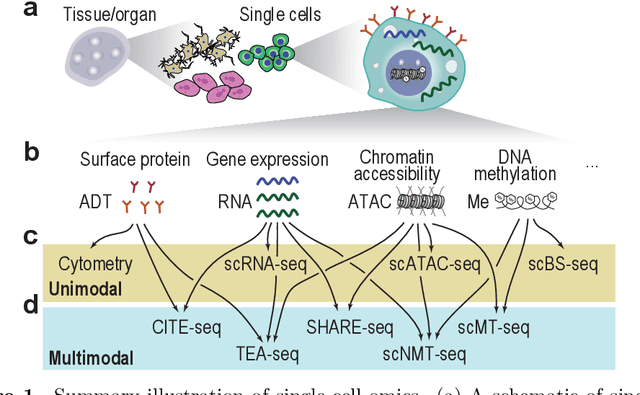
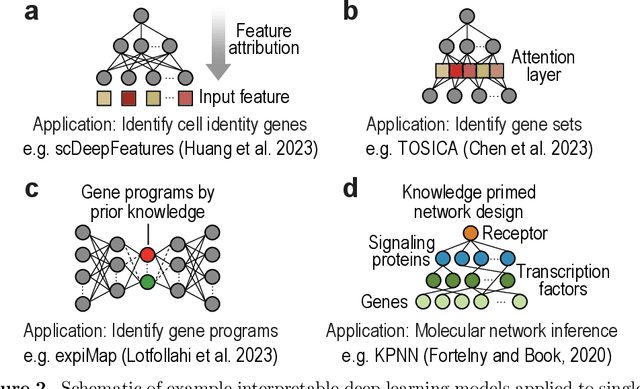
Abstract:Recent developments in single-cell omics technologies have enabled the quantification of molecular profiles in individual cells at an unparalleled resolution. Deep learning, a rapidly evolving sub-field of machine learning, has instilled a significant interest in single-cell omics research due to its remarkable success in analysing heterogeneous high-dimensional single-cell omics data. Nevertheless, the inherent multi-layer nonlinear architecture of deep learning models often makes them `black boxes' as the reasoning behind predictions is often unknown and not transparent to the user. This has stimulated an increasing body of research for addressing the lack of interpretability in deep learning models, especially in single-cell omics data analyses, where the identification and understanding of molecular regulators are crucial for interpreting model predictions and directing downstream experimental validations. In this work, we introduce the basics of single-cell omics technologies and the concept of interpretable deep learning. This is followed by a review of the recent interpretable deep learning models applied to various single-cell omics research. Lastly, we highlight the current limitations and discuss potential future directions. We anticipate this review to bring together the single-cell and machine learning research communities to foster future development and application of interpretable deep learning in single-cell omics research.
Feature selection revisited in the single-cell era
Oct 27, 2021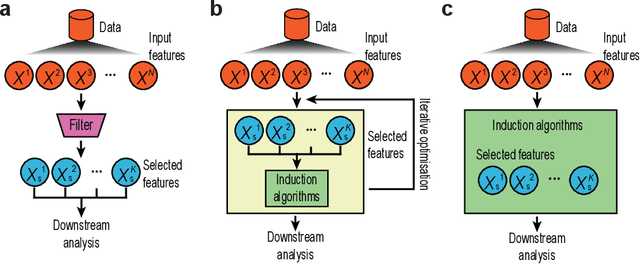
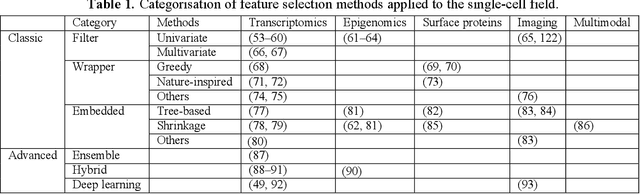
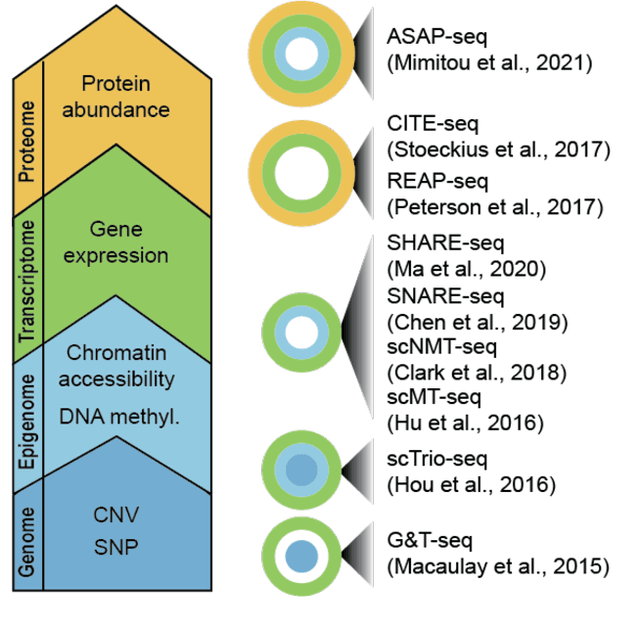
Abstract:Feature selection techniques are essential for high-dimensional data analysis. In the last two decades, their popularity has been fuelled by the increasing availability of high-throughput biomolecular data where high-dimensionality is a common data property. Recent advances in biotechnologies enable global profiling of various molecular and cellular features at single-cell resolution, resulting in large-scale datasets with increased complexity. These technological developments have led to a resurgence in feature selection research and application in the single-cell field. Here, we revisit feature selection techniques and summarise recent developments. We review their versatile application to a range of single-cell data types including those generated from traditional cytometry and imaging technologies and the latest array of single-cell omics technologies. We highlight some of the challenges and future directions on which feature selection could have a significant impact. Finally, we consider the scalability and make general recommendations on the utility of each type of feature selection method. We hope this review serves as a reference point to stimulate future research and application of feature selection in the single-cell era.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge