Theofanis Karaletsos
Parallel Token Prediction for Language Models
Dec 24, 2025Abstract:We propose Parallel Token Prediction (PTP), a universal framework for parallel sequence generation in language models. PTP jointly predicts multiple dependent tokens in a single transformer call by incorporating the sampling procedure into the model. This reduces the latency bottleneck of autoregressive decoding, and avoids the restrictive independence assumptions common in existing multi-token prediction methods. We prove that PTP can represent arbitrary autoregressive sequence distributions. PTP is trained either by distilling an existing model or through inverse autoregressive training without a teacher. Experimentally, we achieve state-of-the-art speculative decoding performance on Vicuna-7B by accepting over four tokens per step on Spec-Bench. The universality of our framework indicates that parallel generation of long sequences is feasible without loss of modeling power.
How Well Do LLMs Understand Drug Mechanisms? A Knowledge + Reasoning Evaluation Dataset
Nov 09, 2025Abstract:Two scientific fields showing increasing interest in pre-trained large language models (LLMs) are drug development / repurposing, and personalized medicine. For both, LLMs have to demonstrate factual knowledge as well as a deep understanding of drug mechanisms, so they can recall and reason about relevant knowledge in novel situations. Drug mechanisms of action are described as a series of interactions between biomedical entities, which interlink into one or more chains directed from the drug to the targeted disease. Composing the effects of the interactions in a candidate chain leads to an inference about whether the drug might be useful or not for that disease. We introduce a dataset that evaluates LLMs on both factual knowledge of known mechanisms, and their ability to reason about them under novel situations, presented as counterfactuals that the models are unlikely to have seen during training. Using this dataset, we show that o4-mini outperforms the 4o, o3, and o3-mini models from OpenAI, and the recent small Qwen3-4B-thinking model closely matches o4-mini's performance, even outperforming it in some cases. We demonstrate that the open world setting for reasoning tasks, which requires the model to recall relevant knowledge, is more challenging than the closed world setting where the needed factual knowledge is provided. We also show that counterfactuals affecting internal links in the reasoning chain present a much harder task than those affecting a link from the drug mentioned in the prompt.
Variational Control for Guidance in Diffusion Models
Feb 06, 2025



Abstract:Diffusion models exhibit excellent sample quality, but existing guidance methods often require additional model training or are limited to specific tasks. We revisit guidance in diffusion models from the perspective of variational inference and control, introducing Diffusion Trajectory Matching (DTM) that enables guiding pretrained diffusion trajectories to satisfy a terminal cost. DTM unifies a broad class of guidance methods and enables novel instantiations. We introduce a new method within this framework that achieves state-of-the-art results on several linear and (blind) non-linear inverse problems without requiring additional model training or modifications. For instance, in ImageNet non-linear deblurring, our model achieves an FID score of 34.31, significantly improving over the best pretrained-method baseline (FID 78.07). We will make the code available in a future update.
Adjusting Pretrained Backbones for Performativity
Oct 06, 2024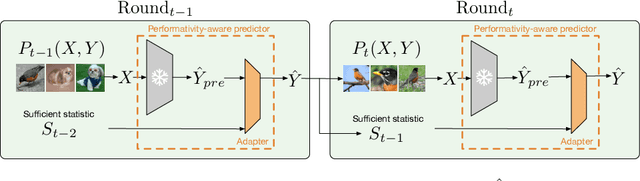



Abstract:With the widespread deployment of deep learning models, they influence their environment in various ways. The induced distribution shifts can lead to unexpected performance degradation in deployed models. Existing methods to anticipate performativity typically incorporate information about the deployed model into the feature vector when predicting future outcomes. While enjoying appealing theoretical properties, modifying the input dimension of the prediction task is often not practical. To address this, we propose a novel technique to adjust pretrained backbones for performativity in a modular way, achieving better sample efficiency and enabling the reuse of existing deep learning assets. Focusing on performative label shift, the key idea is to train a shallow adapter module to perform a Bayes-optimal label shift correction to the backbone's logits given a sufficient statistic of the model to be deployed. As such, our framework decouples the construction of input-specific feature embeddings from the mechanism governing performativity. Motivated by dynamic benchmarking as a use-case, we evaluate our approach under adversarial sampling, for vision and language tasks. We show how it leads to smaller loss along the retraining trajectory and enables us to effectively select among candidate models to anticipate performance degradations. More broadly, our work provides a first baseline for addressing performativity in deep learning.
How to Build the Virtual Cell with Artificial Intelligence: Priorities and Opportunities
Sep 18, 2024

Abstract:The cell is arguably the smallest unit of life and is central to understanding biology. Accurate modeling of cells is important for this understanding as well as for determining the root causes of disease. Recent advances in artificial intelligence (AI), combined with the ability to generate large-scale experimental data, present novel opportunities to model cells. Here we propose a vision of AI-powered Virtual Cells, where robust representations of cells and cellular systems under different conditions are directly learned from growing biological data across measurements and scales. We discuss desired capabilities of AI Virtual Cells, including generating universal representations of biological entities across scales, and facilitating interpretable in silico experiments to predict and understand their behavior using Virtual Instruments. We further address the challenges, opportunities and requirements to realize this vision including data needs, evaluation strategies, and community standards and engagement to ensure biological accuracy and broad utility. We envision a future where AI Virtual Cells help identify new drug targets, predict cellular responses to perturbations, as well as scale hypothesis exploration. With open science collaborations across the biomedical ecosystem that includes academia, philanthropy, and the biopharma and AI industries, a comprehensive predictive understanding of cell mechanisms and interactions is within reach.
Position Paper: Bayesian Deep Learning in the Age of Large-Scale AI
Feb 06, 2024
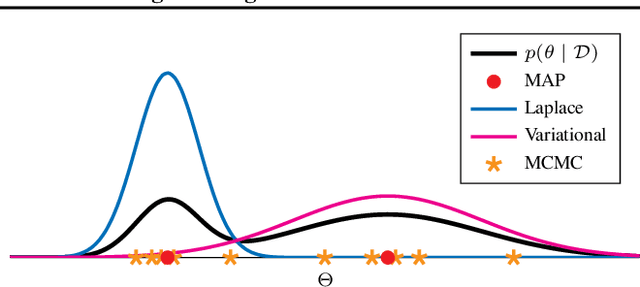
Abstract:In the current landscape of deep learning research, there is a predominant emphasis on achieving high predictive accuracy in supervised tasks involving large image and language datasets. However, a broader perspective reveals a multitude of overlooked metrics, tasks, and data types, such as uncertainty, active and continual learning, and scientific data, that demand attention. Bayesian deep learning (BDL) constitutes a promising avenue, offering advantages across these diverse settings. This paper posits that BDL can elevate the capabilities of deep learning. It revisits the strengths of BDL, acknowledges existing challenges, and highlights some exciting research avenues aimed at addressing these obstacles. Looking ahead, the discussion focuses on possible ways to combine large-scale foundation models with BDL to unlock their full potential.
Modelling Cellular Perturbations with the Sparse Additive Mechanism Shift Variational Autoencoder
Nov 05, 2023



Abstract:Generative models of observations under interventions have been a vibrant topic of interest across machine learning and the sciences in recent years. For example, in drug discovery, there is a need to model the effects of diverse interventions on cells in order to characterize unknown biological mechanisms of action. We propose the Sparse Additive Mechanism Shift Variational Autoencoder, SAMS-VAE, to combine compositionality, disentanglement, and interpretability for perturbation models. SAMS-VAE models the latent state of a perturbed sample as the sum of a local latent variable capturing sample-specific variation and sparse global variables of latent intervention effects. Crucially, SAMS-VAE sparsifies these global latent variables for individual perturbations to identify disentangled, perturbation-specific latent subspaces that are flexibly composable. We evaluate SAMS-VAE both quantitatively and qualitatively on a range of tasks using two popular single cell sequencing datasets. In order to measure perturbation-specific model-properties, we also introduce a framework for evaluation of perturbation models based on average treatment effects with links to posterior predictive checks. SAMS-VAE outperforms comparable models in terms of generalization across in-distribution and out-of-distribution tasks, including a combinatorial reasoning task under resource paucity, and yields interpretable latent structures which correlate strongly to known biological mechanisms. Our results suggest SAMS-VAE is an interesting addition to the modeling toolkit for machine learning-driven scientific discovery.
Compositional Deep Probabilistic Models of DNA Encoded Libraries
Oct 20, 2023Abstract:DNA-Encoded Library (DEL) has proven to be a powerful tool that utilizes combinatorially constructed small molecules to facilitate highly-efficient screening assays. These selection experiments, involving multiple stages of washing, elution, and identification of potent binders via unique DNA barcodes, often generate complex data. This complexity can potentially mask the underlying signals, necessitating the application of computational tools such as machine learning to uncover valuable insights. We introduce a compositional deep probabilistic model of DEL data, DEL-Compose, which decomposes molecular representations into their mono-synthon, di-synthon, and tri-synthon building blocks and capitalizes on the inherent hierarchical structure of these molecules by modeling latent reactions between embedded synthons. Additionally, we investigate methods to improve the observation models for DEL count data such as integrating covariate factors to more effectively account for data noise. Across two popular public benchmark datasets (CA-IX and HRP), our model demonstrates strong performance compared to count baselines, enriches the correct pharmacophores, and offers valuable insights via its intrinsic interpretable structure, thereby providing a robust tool for the analysis of DEL data.
Channel Vision Transformers: An Image Is Worth C x 16 x 16 Words
Oct 13, 2023Abstract:Vision Transformer (ViT) has emerged as a powerful architecture in the realm of modern computer vision. However, its application in certain imaging fields, such as microscopy and satellite imaging, presents unique challenges. In these domains, images often contain multiple channels, each carrying semantically distinct and independent information. Furthermore, the model must demonstrate robustness to sparsity in input channels, as they may not be densely available during training or testing. In this paper, we propose a modification to the ViT architecture that enhances reasoning across the input channels and introduce Hierarchical Channel Sampling (HCS) as an additional regularization technique to ensure robustness when only partial channels are presented during test time. Our proposed model, ChannelViT, constructs patch tokens independently from each input channel and utilizes a learnable channel embedding that is added to the patch tokens, similar to positional embeddings. We evaluate the performance of ChannelViT on ImageNet, JUMP-CP (microscopy cell imaging), and So2Sat (satellite imaging). Our results show that ChannelViT outperforms ViT on classification tasks and generalizes well, even when a subset of input channels is used during testing. Across our experiments, HCS proves to be a powerful regularizer, independent of the architecture employed, suggesting itself as a straightforward technique for robust ViT training. Lastly, we find that ChannelViT generalizes effectively even when there is limited access to all channels during training, highlighting its potential for multi-channel imaging under real-world conditions with sparse sensors. Our code is available at https://github.com/insitro/ChannelViT.
Contextual Vision Transformers for Robust Representation Learning
May 30, 2023



Abstract:We present Contextual Vision Transformers (ContextViT), a method for producing robust feature representations for images exhibiting grouped structure such as covariates. ContextViT introduces an extra context token to encode group-specific information, allowing the model to explain away group-specific covariate structures while keeping core visual features shared across groups. Specifically, given an input image, Context-ViT maps images that share the same covariate into this context token appended to the input image tokens to capture the effects of conditioning the model on group membership. We furthermore introduce a context inference network to predict such tokens on the fly given a few samples from a group distribution, enabling ContextViT to generalize to new testing distributions at inference time. We illustrate the performance of ContextViT through a diverse range of applications. In supervised fine-tuning, we demonstrate that augmenting pre-trained ViTs with additional context conditioning leads to significant improvements in out-of-distribution generalization on iWildCam and FMoW. We also explored self-supervised representation learning with ContextViT. Our experiments on the Camelyon17 pathology imaging benchmark and the cpg-0000 microscopy imaging benchmark demonstrate that ContextViT excels in learning stable image featurizations amidst covariate shift, consistently outperforming its ViT counterpart.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge