Nikolas Lessmann
Semi-Supervised Segmentation via Embedding Matching
Jul 05, 2024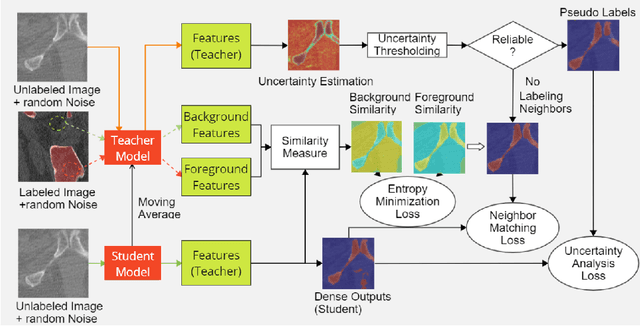
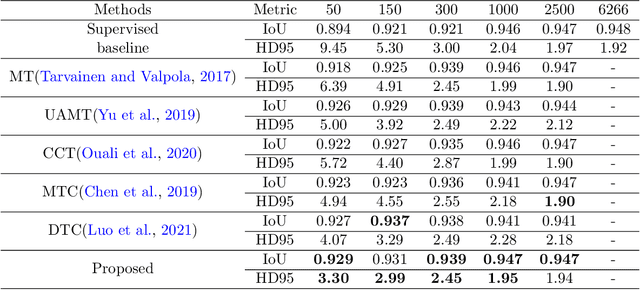
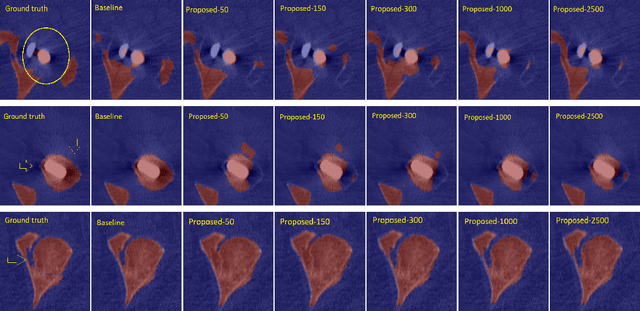

Abstract:Deep convolutional neural networks are widely used in medical image segmentation but require many labeled images for training. Annotating three-dimensional medical images is a time-consuming and costly process. To overcome this limitation, we propose a novel semi-supervised segmentation method that leverages mostly unlabeled images and a small set of labeled images in training. Our approach involves assessing prediction uncertainty to identify reliable predictions on unlabeled voxels from the teacher model. These voxels serve as pseudo-labels for training the student model. In voxels where the teacher model produces unreliable predictions, pseudo-labeling is carried out based on voxel-wise embedding correspondence using reference voxels from labeled images. We applied this method to automate hip bone segmentation in CT images, achieving notable results with just 4 CT scans. The proposed approach yielded a Hausdorff distance with 95th percentile (HD95) of 3.30 and IoU of 0.929, surpassing existing methods achieving HD95 (4.07) and IoU (0.927) at their best.
Transfer learning from a sparsely annotated dataset of 3D medical images
Nov 08, 2023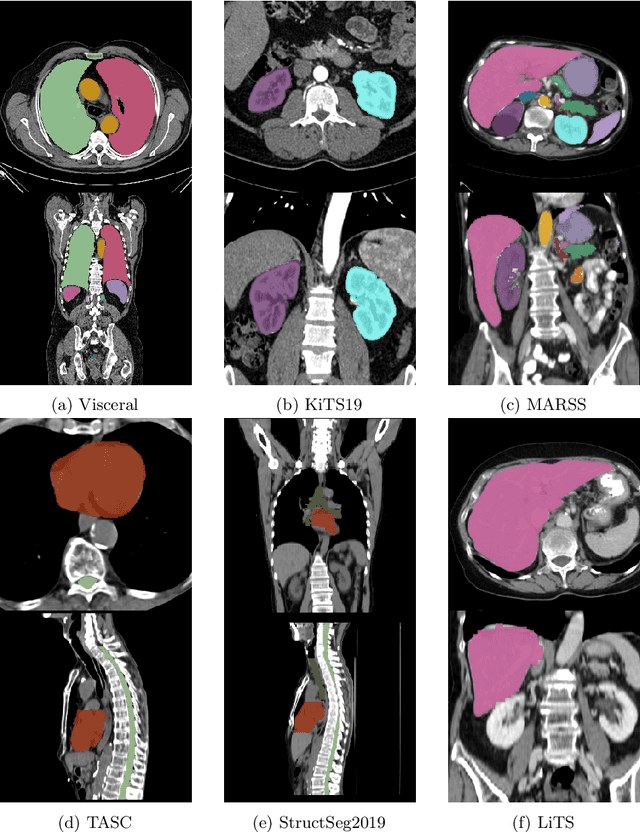
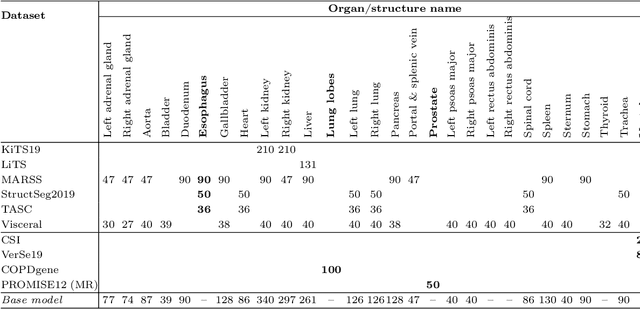
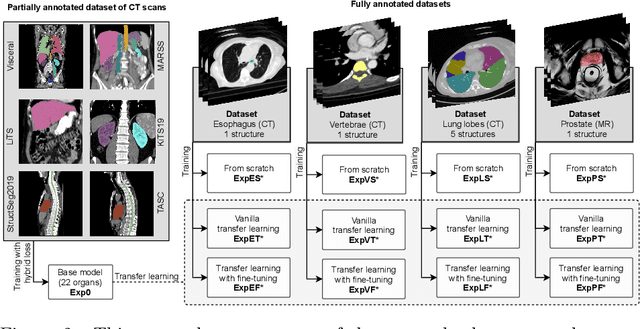
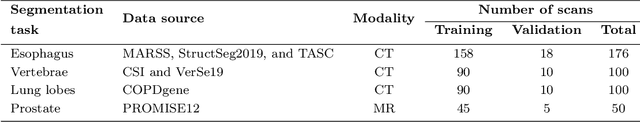
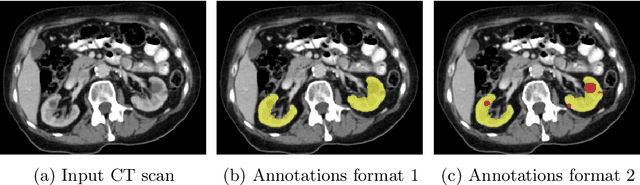
Abstract:Transfer learning leverages pre-trained model features from a large dataset to save time and resources when training new models for various tasks, potentially enhancing performance. Due to the lack of large datasets in the medical imaging domain, transfer learning from one medical imaging model to other medical imaging models has not been widely explored. This study explores the use of transfer learning to improve the performance of deep convolutional neural networks for organ segmentation in medical imaging. A base segmentation model (3D U-Net) was trained on a large and sparsely annotated dataset; its weights were used for transfer learning on four new down-stream segmentation tasks for which a fully annotated dataset was available. We analyzed the training set size's influence to simulate scarce data. The results showed that transfer learning from the base model was beneficial when small datasets were available, providing significant performance improvements; where fine-tuning the base model is more beneficial than updating all the network weights with vanilla transfer learning. Transfer learning with fine-tuning increased the performance by up to 0.129 (+28\%) Dice score than experiments trained from scratch, and on average 23 experiments increased the performance by 0.029 Dice score in the new segmentation tasks. The study also showed that cross-modality transfer learning using CT scans was beneficial. The findings of this study demonstrate the potential of transfer learning to improve the efficiency of annotation and increase the accessibility of accurate organ segmentation in medical imaging, ultimately leading to improved patient care. We made the network definition and weights publicly available to benefit other users and researchers.
Kidney abnormality segmentation in thorax-abdomen CT scans
Sep 06, 2023


Abstract:In this study, we introduce a deep learning approach for segmenting kidney parenchyma and kidney abnormalities to support clinicians in identifying and quantifying renal abnormalities such as cysts, lesions, masses, metastases, and primary tumors. Our end-to-end segmentation method was trained on 215 contrast-enhanced thoracic-abdominal CT scans, with half of these scans containing one or more abnormalities. We began by implementing our own version of the original 3D U-Net network and incorporated four additional components: an end-to-end multi-resolution approach, a set of task-specific data augmentations, a modified loss function using top-$k$, and spatial dropout. Furthermore, we devised a tailored post-processing strategy. Ablation studies demonstrated that each of the four modifications enhanced kidney abnormality segmentation performance, while three out of four improved kidney parenchyma segmentation. Subsequently, we trained the nnUNet framework on our dataset. By ensembling the optimized 3D U-Net and the nnUNet with our specialized post-processing, we achieved marginally superior results. Our best-performing model attained Dice scores of 0.965 and 0.947 for segmenting kidney parenchyma in two test sets (20 scans without abnormalities and 30 with abnormalities), outperforming an independent human observer who scored 0.944 and 0.925, respectively. In segmenting kidney abnormalities within the 30 test scans containing them, the top-performing method achieved a Dice score of 0.585, while an independent second human observer reached a score of 0.664, suggesting potential for further improvement in computerized methods. All training data is available to the research community under a CC-BY 4.0 license on https://doi.org/10.5281/zenodo.8014289
Lumbar spine segmentation in MR images: a dataset and a public benchmark
Jun 22, 2023Abstract:This paper presents a large publicly available multi-center lumbar spine magnetic resonance imaging (MRI) dataset with reference segmentations of vertebrae, intervertebral discs (IVDs), and spinal canal. The dataset includes 447 sagittal T1 and T2 MRI series from 218 patients with a history of low back pain. It was collected from four different hospitals and was divided into a training (179 patients) and validation (39 patients) set. An iterative data annotation approach was used by training a segmentation algorithm on a small part of the dataset, enabling semi-automatic segmentation of the remaining images. The algorithm provided an initial segmentation, which was subsequently reviewed, manually corrected, and added to the training data. We provide reference performance values for this baseline algorithm and nnU-Net, which performed comparably. We set up a continuous segmentation challenge to allow for a fair comparison of different segmentation algorithms. This study may encourage wider collaboration in the field of spine segmentation, and improve the diagnostic value of lumbar spine MRI.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021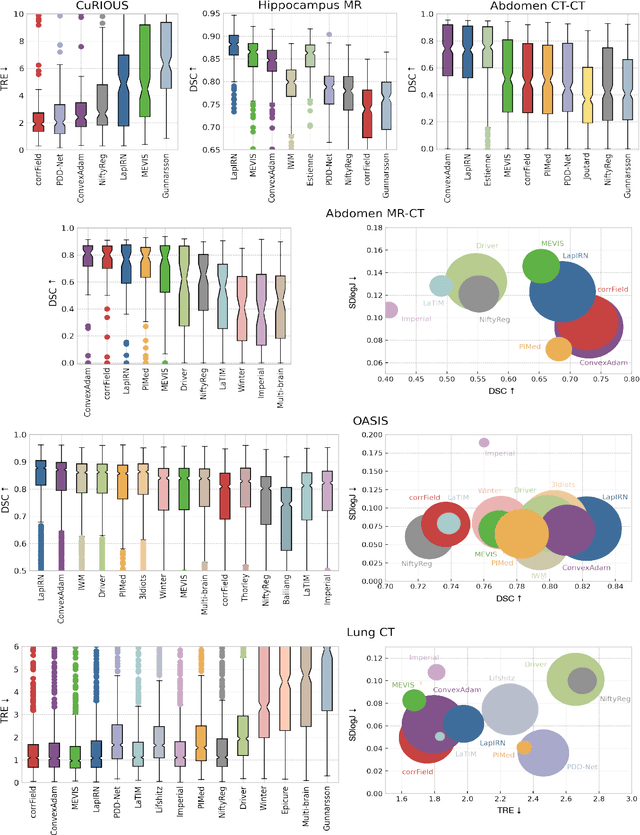
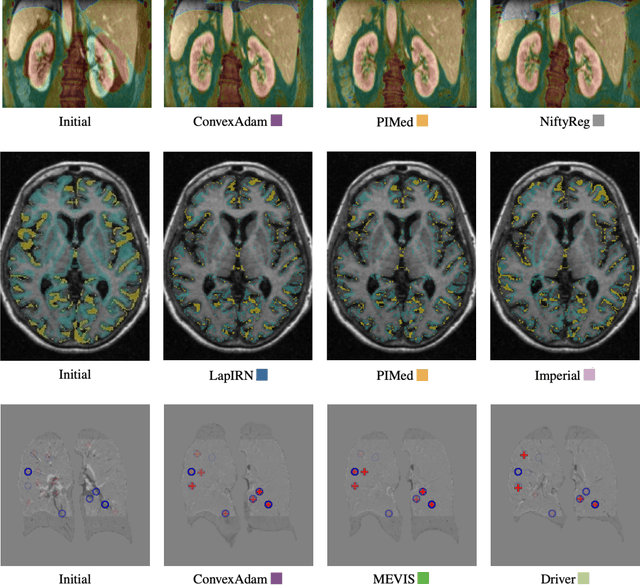
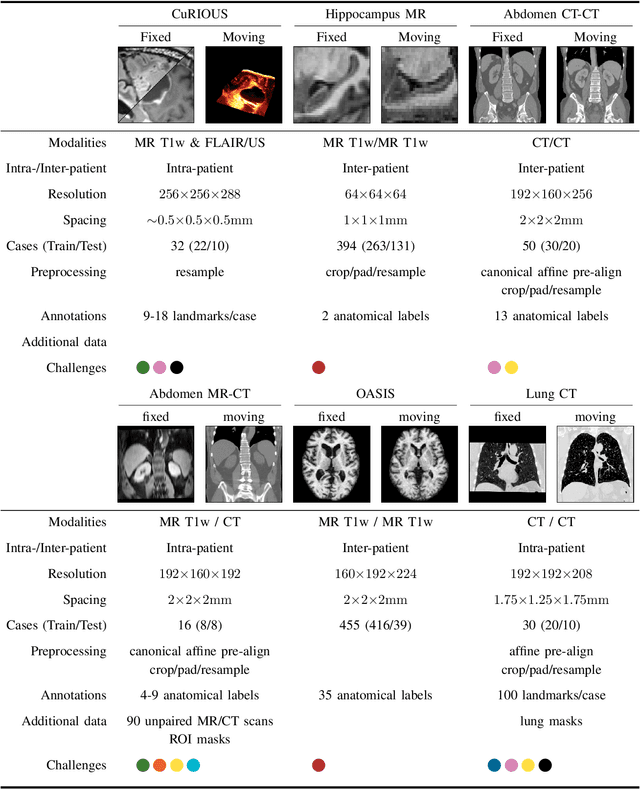
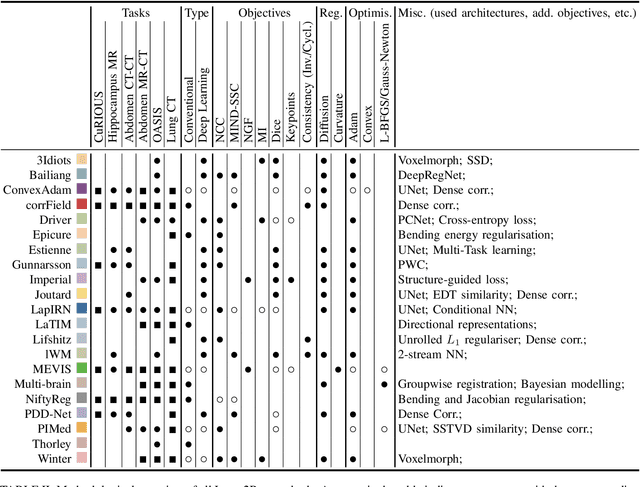
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Constraining Volume Change in Learned Image Registration for Lung CTs
Nov 29, 2020



Abstract:Deep-learning-based registration methods emerged as a fast alternative to conventional registration methods. However, these methods often still cannot achieve the same performance as conventional registration methods, because they are either limited to small deformation or they fail to handle a superposition of large and small deformations without producing implausible deformation fields with foldings inside. In this paper, we identify important strategies of conventional registration methods for lung registration and successfully developed the deep-learning counterpart. We employ a Gaussian-pyramid-based multilevel framework that can solve the image registration optimization in a coarse-to-fine fashion. Furthermore, we prevent foldings of the deformation field and restrict the determinant of the Jacobian to physiologically meaningful values by combining a volume change penalty with a curvature regularizer in the loss function. Keypoint correspondences are integrated to focus on the alignment of smaller structures. We perform an extensive evaluation to assess the accuracy, the robustness, the plausibility of the estimated deformation fields, and the transferability of our registration approach. We show that it archives state-of-the-art results on the COPDGene dataset compared to the challenge winning conventional registration method with much shorter execution time.
Improving Automated COVID-19 Grading with Convolutional Neural Networks in Computed Tomography Scans: An Ablation Study
Sep 21, 2020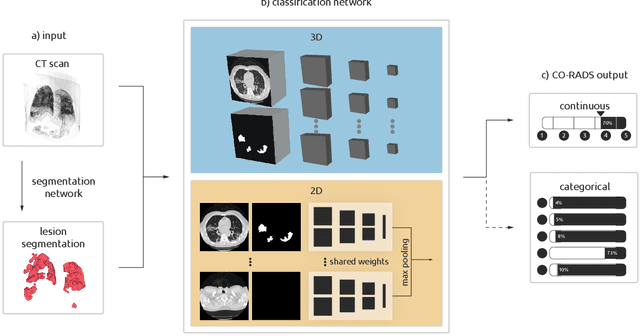
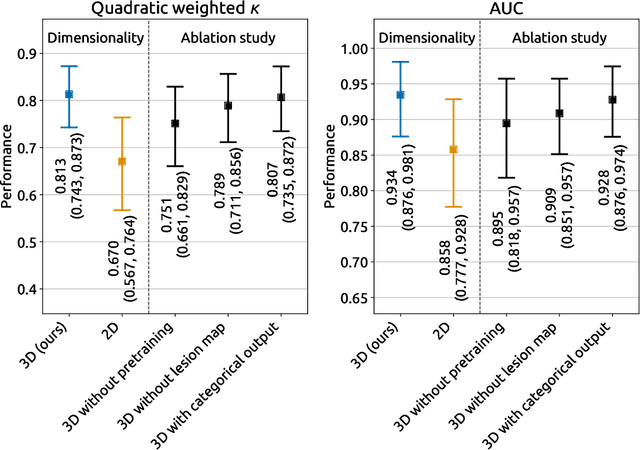
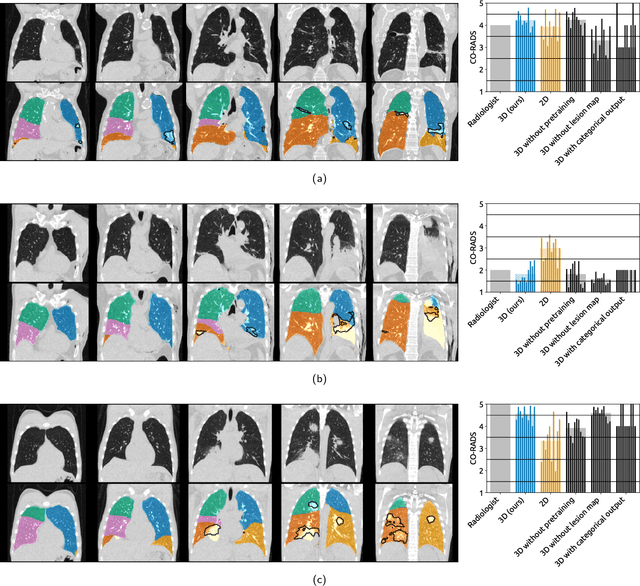
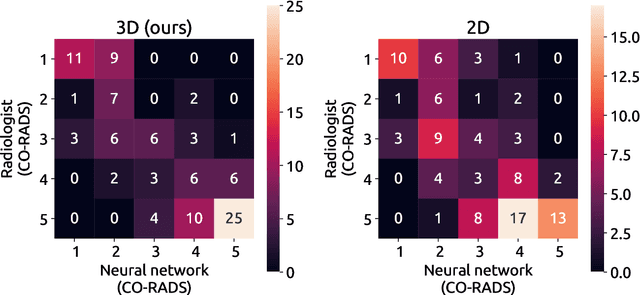
Abstract:Amidst the ongoing pandemic, several studies have shown that COVID-19 classification and grading using computed tomography (CT) images can be automated with convolutional neural networks (CNNs). Many of these studies focused on reporting initial results of algorithms that were assembled from commonly used components. The choice of these components was often pragmatic rather than systematic. For instance, several studies used 2D CNNs even though these might not be optimal for handling 3D CT volumes. This paper identifies a variety of components that increase the performance of CNN-based algorithms for COVID-19 grading from CT images. We investigated the effectiveness of using a 3D CNN instead of a 2D CNN, of using transfer learning to initialize the network, of providing automatically computed lesion maps as additional network input, and of predicting a continuous instead of a categorical output. A 3D CNN with these components achieved an area under the ROC curve (AUC) of 0.934 on our test set of 105 CT scans and an AUC of 0.923 on a publicly available set of 742 CT scans, a substantial improvement in comparison with a previously published 2D CNN. An ablation study demonstrated that in addition to using a 3D CNN instead of a 2D CNN transfer learning contributed the most and continuous output contributed the least to improving the model performance.
Random smooth gray value transformations for cross modality learning with gray value invariant networks
Mar 13, 2020

Abstract:Random transformations are commonly used for augmentation of the training data with the goal of reducing the uniformity of the training samples. These transformations normally aim at variations that can be expected in images from the same modality. Here, we propose a simple method for transforming the gray values of an image with the goal of reducing cross modality differences. This approach enables segmentation of the lumbar vertebral bodies in CT images using a network trained exclusively with MR images. The source code is made available at https://github.com/nlessmann/rsgt
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020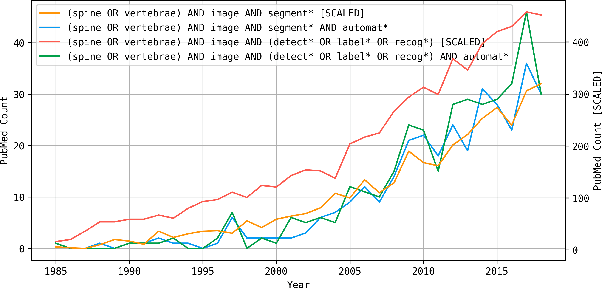
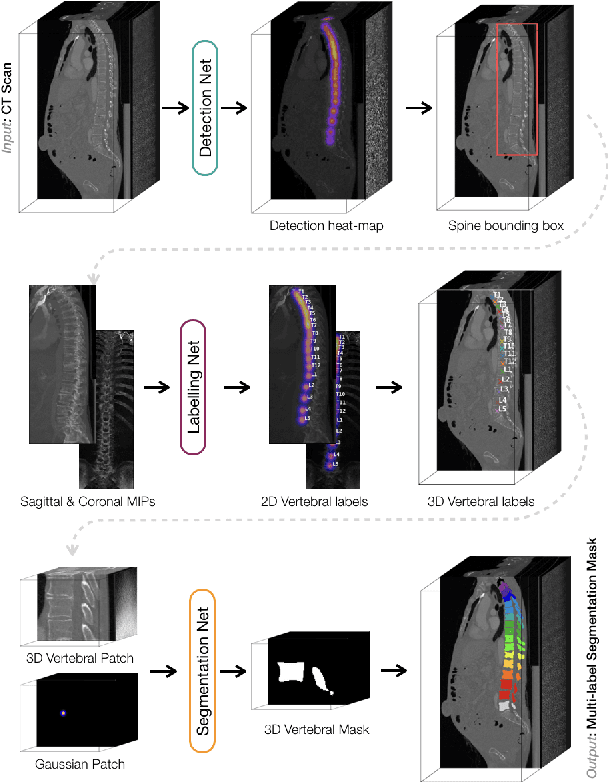
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
Direct Automatic Coronary Calcium Scoring in Cardiac and Chest CT
Feb 12, 2019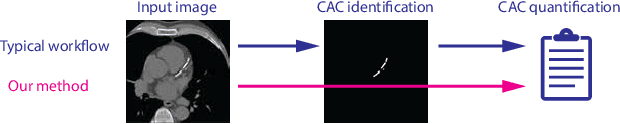
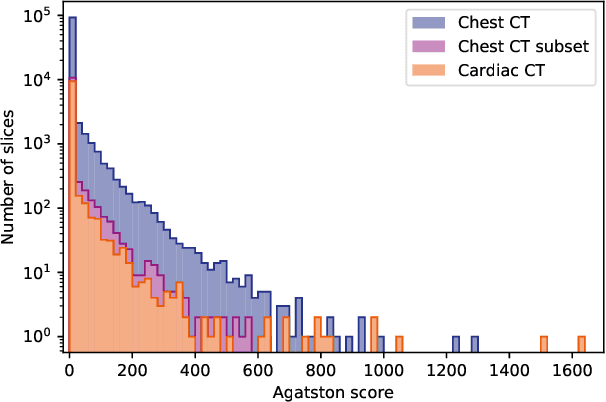
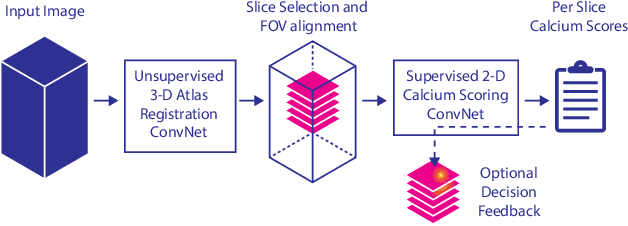
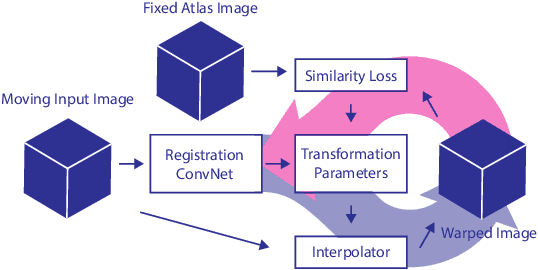
Abstract:Cardiovascular disease (CVD) is the global leading cause of death. A strong risk factor for CVD events is the amount of coronary artery calcium (CAC). To meet demands of the increasing interest in quantification of CAC, i.e. coronary calcium scoring, especially as an unrequested finding for screening and research, automatic methods have been proposed. Current automatic calcium scoring methods are relatively computationally expensive and only provide scores for one type of CT. To address this, we propose a computationally efficient method that employs two ConvNets: the first performs registration to align the fields of view of input CTs and the second performs direct regression of the calcium score, thereby circumventing time-consuming intermediate CAC segmentation. Optional decision feedback provides insight in the regions that contributed to the calcium score. Experiments were performed using 903 cardiac CT and 1,687 chest CT scans. The method predicted calcium scores in less than 0.3 s. Intra-class correlation coefficient between predicted and manual calcium scores was 0.98 for both cardiac and chest CT. The method showed almost perfect agreement between automatic and manual CVD risk categorization in both datasets, with a linearly weighted Cohen's kappa of 0.95 in cardiac CT and 0.93 in chest CT. Performance is similar to that of state-of-the-art methods, but the proposed method is hundreds of times faster. By providing visual feedback, insight is given in the decision process, making it readily implementable in clinical and research settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge