Colin Jacobs
Fairness Evaluation of Risk Estimation Models for Lung Cancer Screening
Dec 23, 2025Abstract:Lung cancer is the leading cause of cancer-related mortality in adults worldwide. Screening high-risk individuals with annual low-dose CT (LDCT) can support earlier detection and reduce deaths, but widespread implementation may strain the already limited radiology workforce. AI models have shown potential in estimating lung cancer risk from LDCT scans. However, high-risk populations for lung cancer are diverse, and these models' performance across demographic groups remains an open question. In this study, we drew on the considerations on confounding factors and ethically significant biases outlined in the JustEFAB framework to evaluate potential performance disparities and fairness in two deep learning risk estimation models for lung cancer screening: the Sybil lung cancer risk model and the Venkadesh21 nodule risk estimator. We also examined disparities in the PanCan2b logistic regression model recommended in the British Thoracic Society nodule management guideline. Both deep learning models were trained on data from the US-based National Lung Screening Trial (NLST), and assessed on a held-out NLST validation set. We evaluated AUROC, sensitivity, and specificity across demographic subgroups, and explored potential confounding from clinical risk factors. We observed a statistically significant AUROC difference in Sybil's performance between women (0.88, 95% CI: 0.86, 0.90) and men (0.81, 95% CI: 0.78, 0.84, p < .001). At 90% specificity, Venkadesh21 showed lower sensitivity for Black (0.39, 95% CI: 0.23, 0.59) than White participants (0.69, 95% CI: 0.65, 0.73). These differences were not explained by available clinical confounders and thus may be classified as unfair biases according to JustEFAB. Our findings highlight the importance of improving and monitoring model performance across underrepresented subgroups, and further research on algorithmic fairness, in lung cancer screening.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:025
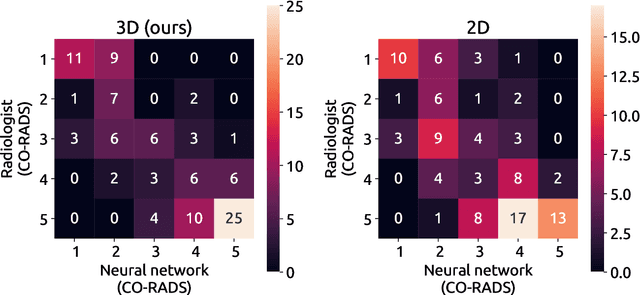
Nodule detection and generation on chest X-rays: NODE21 Challenge
Jan 04, 2024



Abstract:Pulmonary nodules may be an early manifestation of lung cancer, the leading cause of cancer-related deaths among both men and women. Numerous studies have established that deep learning methods can yield high-performance levels in the detection of lung nodules in chest X-rays. However, the lack of gold-standard public datasets slows down the progression of the research and prevents benchmarking of methods for this task. To address this, we organized a public research challenge, NODE21, aimed at the detection and generation of lung nodules in chest X-rays. While the detection track assesses state-of-the-art nodule detection systems, the generation track determines the utility of nodule generation algorithms to augment training data and hence improve the performance of the detection systems. This paper summarizes the results of the NODE21 challenge and performs extensive additional experiments to examine the impact of the synthetically generated nodule training images on the detection algorithm performance.
Transfer learning from a sparsely annotated dataset of 3D medical images
Nov 08, 2023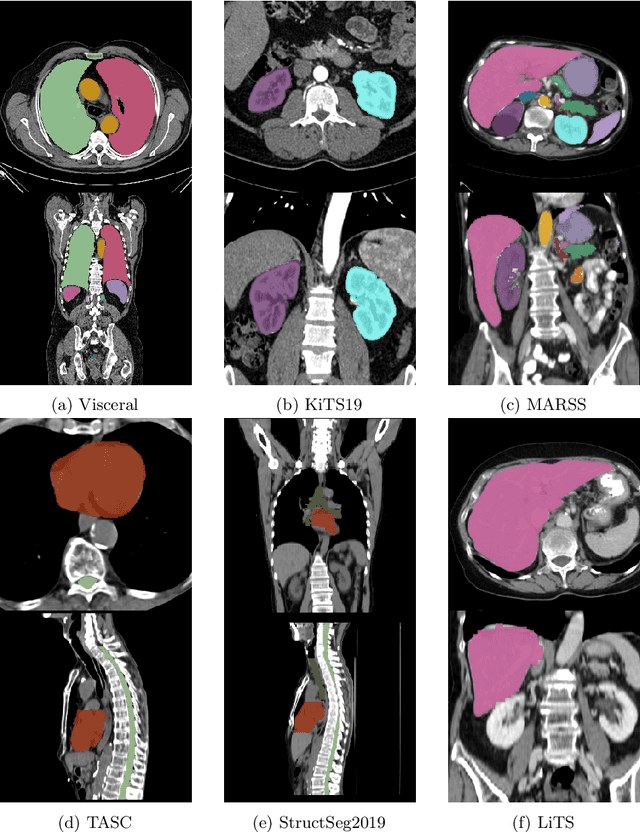
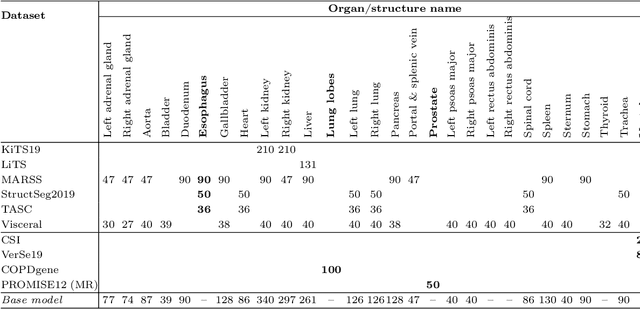
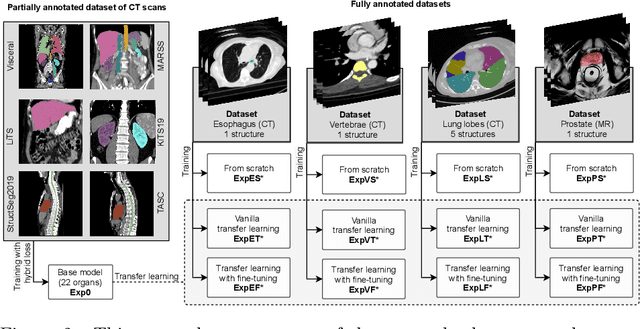
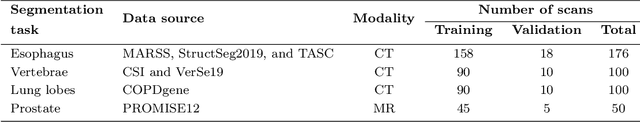
Abstract:Transfer learning leverages pre-trained model features from a large dataset to save time and resources when training new models for various tasks, potentially enhancing performance. Due to the lack of large datasets in the medical imaging domain, transfer learning from one medical imaging model to other medical imaging models has not been widely explored. This study explores the use of transfer learning to improve the performance of deep convolutional neural networks for organ segmentation in medical imaging. A base segmentation model (3D U-Net) was trained on a large and sparsely annotated dataset; its weights were used for transfer learning on four new down-stream segmentation tasks for which a fully annotated dataset was available. We analyzed the training set size's influence to simulate scarce data. The results showed that transfer learning from the base model was beneficial when small datasets were available, providing significant performance improvements; where fine-tuning the base model is more beneficial than updating all the network weights with vanilla transfer learning. Transfer learning with fine-tuning increased the performance by up to 0.129 (+28\%) Dice score than experiments trained from scratch, and on average 23 experiments increased the performance by 0.029 Dice score in the new segmentation tasks. The study also showed that cross-modality transfer learning using CT scans was beneficial. The findings of this study demonstrate the potential of transfer learning to improve the efficiency of annotation and increase the accessibility of accurate organ segmentation in medical imaging, ultimately leading to improved patient care. We made the network definition and weights publicly available to benefit other users and researchers.
Kidney abnormality segmentation in thorax-abdomen CT scans
Sep 06, 2023
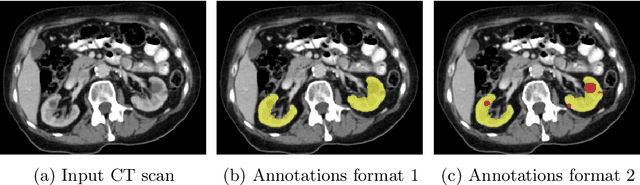


Abstract:In this study, we introduce a deep learning approach for segmenting kidney parenchyma and kidney abnormalities to support clinicians in identifying and quantifying renal abnormalities such as cysts, lesions, masses, metastases, and primary tumors. Our end-to-end segmentation method was trained on 215 contrast-enhanced thoracic-abdominal CT scans, with half of these scans containing one or more abnormalities. We began by implementing our own version of the original 3D U-Net network and incorporated four additional components: an end-to-end multi-resolution approach, a set of task-specific data augmentations, a modified loss function using top-$k$, and spatial dropout. Furthermore, we devised a tailored post-processing strategy. Ablation studies demonstrated that each of the four modifications enhanced kidney abnormality segmentation performance, while three out of four improved kidney parenchyma segmentation. Subsequently, we trained the nnUNet framework on our dataset. By ensembling the optimized 3D U-Net and the nnUNet with our specialized post-processing, we achieved marginally superior results. Our best-performing model attained Dice scores of 0.965 and 0.947 for segmenting kidney parenchyma in two test sets (20 scans without abnormalities and 30 with abnormalities), outperforming an independent human observer who scored 0.944 and 0.925, respectively. In segmenting kidney abnormalities within the 30 test scans containing them, the top-performing method achieved a Dice score of 0.585, while an independent second human observer reached a score of 0.664, suggesting potential for further improvement in computerized methods. All training data is available to the research community under a CC-BY 4.0 license on https://doi.org/10.5281/zenodo.8014289
Emphysema Subtyping on Thoracic Computed Tomography Scans using Deep Neural Networks
Sep 05, 2023Abstract:Accurate identification of emphysema subtypes and severity is crucial for effective management of COPD and the study of disease heterogeneity. Manual analysis of emphysema subtypes and severity is laborious and subjective. To address this challenge, we present a deep learning-based approach for automating the Fleischner Society's visual score system for emphysema subtyping and severity analysis. We trained and evaluated our algorithm using 9650 subjects from the COPDGene study. Our algorithm achieved the predictive accuracy at 52\%, outperforming a previously published method's accuracy of 45\%. In addition, the agreement between the predicted scores of our method and the visual scores was good, where the previous method obtained only moderate agreement. Our approach employs a regression training strategy to generate categorical labels while simultaneously producing high-resolution localized activation maps for visualizing the network predictions. By leveraging these dense activation maps, our method possesses the capability to compute the percentage of emphysema involvement per lung in addition to categorical severity scores. Furthermore, the proposed method extends its predictive capabilities beyond centrilobular emphysema to include paraseptal emphysema subtypes.
Structure and position-aware graph neural network for airway labeling
Jan 12, 2022



Abstract:We present a novel graph-based approach for labeling the anatomical branches of a given airway tree segmentation. The proposed method formulates airway labeling as a branch classification problem in the airway tree graph, where branch features are extracted using convolutional neural networks (CNN) and enriched using graph neural networks. Our graph neural network is structure-aware by having each node aggregate information from its local neighbors and position-aware by encoding node positions in the graph. We evaluated the proposed method on 220 airway trees from subjects with various severity stages of Chronic Obstructive Pulmonary Disease (COPD). The results demonstrate that our approach is computationally efficient and significantly improves branch classification performance than the baseline method. The overall average accuracy of our method reaches 91.18\% for labeling all 18 segmental airway branches, compared to 83.83\% obtained by the standard CNN method. We published our source code at https://github.com/DIAGNijmegen/spgnn. The proposed algorithm is also publicly available at https://grand-challenge.org/algorithms/airway-anatomical-labeling/.
Deep Clustering Activation Maps for Emphysema Subtyping
Jun 01, 2021

Abstract:We propose a deep learning clustering method that exploits dense features from a segmentation network for emphysema subtyping from computed tomography (CT) scans. Using dense features enables high-resolution visualization of image regions corresponding to the cluster assignment via dense clustering activation maps (dCAMs). This approach provides model interpretability. We evaluated clustering results on 500 subjects from the COPDGenestudy, where radiologists manually annotated emphysema sub-types according to their visual CT assessment. We achieved a 43% unsupervised clustering accuracy, outperforming our baseline at 41% and yielding results comparable to supervised classification at 45%. The proposed method also offers a better cluster formation than the baseline, achieving0.54 in silhouette coefficient and 0.55 in David-Bouldin scores.
Dense Regression Activation Maps For Lesion Segmentation in CT scans of COVID-19 patients
May 25, 2021



Abstract:Automatic lesion segmentation on thoracic CT enables rapid quantitative analysis of lung involvement in COVID- 19 infections. Obtaining voxel-level annotations for training segmentation networks is prohibitively expensive. Therefore we propose a weakly-supervised segmentation method based on dense regression activation maps (dRAM). Most advanced weakly supervised segmentation approaches exploit class activation maps (CAMs) to localize objects generated from high-level semantic features at a coarse resolution. As a result, CAMs provide coarse outlines that do not align precisely with the object segmentations. Instead, we exploit dense features from a segmentation network to compute dense regression activation maps (dRAMs) for preserving local details. During training, dRAMs are pooled lobe-wise to regress the per-lobe lesion percentage. In such a way, the network achieves additional information regarding the lesion quantification in comparison with the classification approach. Furthermore, we refine dRAMs based on an attention module and dense conditional random field trained together with the main regression task. The refined dRAMs are served as the pseudo labels for training a final segmentation network. When evaluated on 69 CT scans, our method substantially improves the intersection over union from 0.335 in the CAM-based weakly supervised segmentation method to 0.495.
Improving Automated COVID-19 Grading with Convolutional Neural Networks in Computed Tomography Scans: An Ablation Study
Sep 21, 2020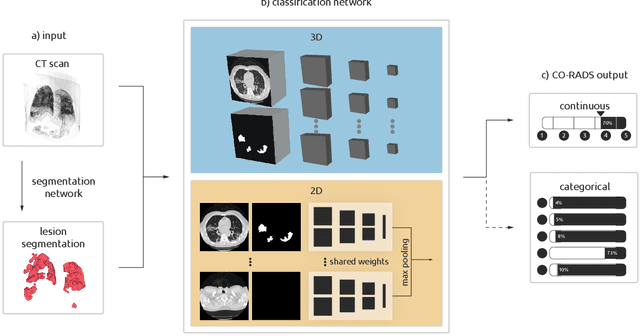
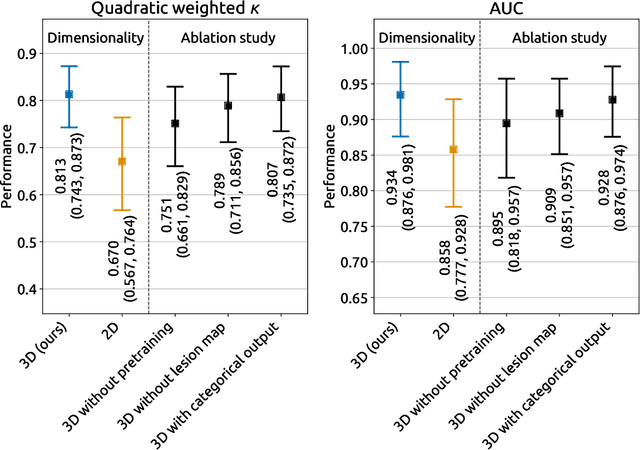
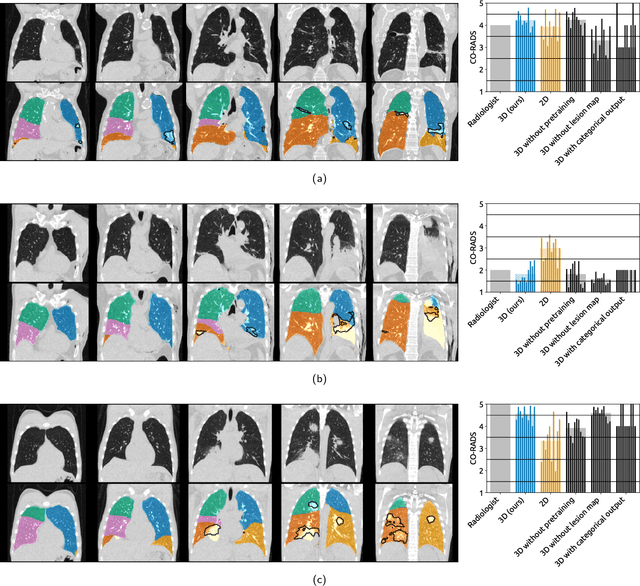
Abstract:Amidst the ongoing pandemic, several studies have shown that COVID-19 classification and grading using computed tomography (CT) images can be automated with convolutional neural networks (CNNs). Many of these studies focused on reporting initial results of algorithms that were assembled from commonly used components. The choice of these components was often pragmatic rather than systematic. For instance, several studies used 2D CNNs even though these might not be optimal for handling 3D CT volumes. This paper identifies a variety of components that increase the performance of CNN-based algorithms for COVID-19 grading from CT images. We investigated the effectiveness of using a 3D CNN instead of a 2D CNN, of using transfer learning to initialize the network, of providing automatically computed lesion maps as additional network input, and of predicting a continuous instead of a categorical output. A 3D CNN with these components achieved an area under the ROC curve (AUC) of 0.934 on our test set of 105 CT scans and an AUC of 0.923 on a publicly available set of 742 CT scans, a substantial improvement in comparison with a previously published 2D CNN. An ablation study demonstrated that in addition to using a 3D CNN instead of a 2D CNN transfer learning contributed the most and continuous output contributed the least to improving the model performance.
Relational Modeling for Robust and Efficient Pulmonary Lobe Segmentation in CT Scans
May 12, 2020



Abstract:Pulmonary lobe segmentation in computed tomography scans is essential for regional assessment of pulmonary diseases. Recent works based on convolution neural networks have achieved good performance for this task. However, they are still limited in capturing structured relationships due to the nature of convolution. The shape of the pulmonary lobes affect each other and their borders relate to the appearance of other structures, such as vessels, airways, and the pleural wall. We argue that such structural relationships play a critical role in the accurate delineation of pulmonary lobes when the lungs are affected by diseases such as COVID-19 or COPD. In this paper, we propose a relational approach (RTSU-Net) that leverages structured relationships by introducing a novel non-local neural network module. The proposed module learns both visual and geometric relationships among all convolution features to produce self-attention weights. With a limited amount of training data available from COVID-19 subjects, we initially train and validate RTSU-Net on a cohort of 5000 subjects from the COPDGene study (4000 for training and 1000 for evaluation). Using models pre-trained on COPDGene, we apply transfer learning to retrain and evaluate RTSU-Net on 470 COVID-19 suspects (370 for retraining and 100 for evaluation). Experimental results show that RTSU-Net outperforms three baselines and performs robustly on cases with severe lung infection due to COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge