Weiyi Xie
SAM Fewshot Finetuning for Anatomical Segmentation in Medical Images
Jul 05, 2024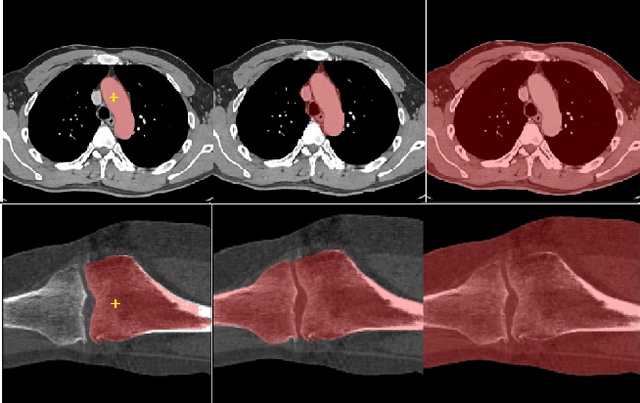
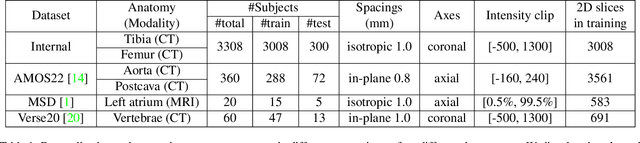
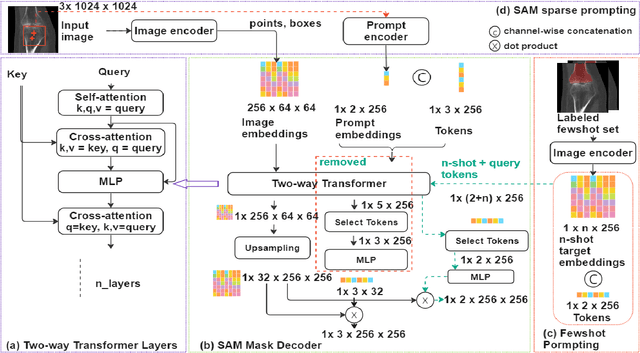
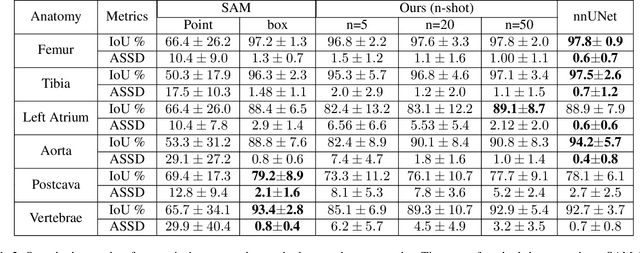
Abstract:We propose a straightforward yet highly effective few-shot fine-tuning strategy for adapting the Segment Anything (SAM) to anatomical segmentation tasks in medical images. Our novel approach revolves around reformulating the mask decoder within SAM, leveraging few-shot embeddings derived from a limited set of labeled images (few-shot collection) as prompts for querying anatomical objects captured in image embeddings. This innovative reformulation greatly reduces the need for time-consuming online user interactions for labeling volumetric images, such as exhaustively marking points and bounding boxes to provide prompts slice by slice. With our method, users can manually segment a few 2D slices offline, and the embeddings of these annotated image regions serve as effective prompts for online segmentation tasks. Our method prioritizes the efficiency of the fine-tuning process by exclusively training the mask decoder through caching mechanisms while keeping the image encoder frozen. Importantly, this approach is not limited to volumetric medical images, but can generically be applied to any 2D/3D segmentation task. To thoroughly evaluate our method, we conducted extensive validation on four datasets, covering six anatomical segmentation tasks across two modalities. Furthermore, we conducted a comparative analysis of different prompting options within SAM and the fully-supervised nnU-Net. The results demonstrate the superior performance of our method compared to SAM employing only point prompts (approximately 50% improvement in IoU) and performs on-par with fully supervised methods whilst reducing the requirement of labeled data by at least an order of magnitude.
Semi-Supervised Segmentation via Embedding Matching
Jul 05, 2024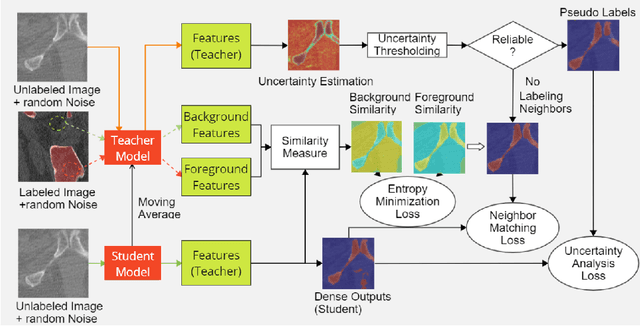
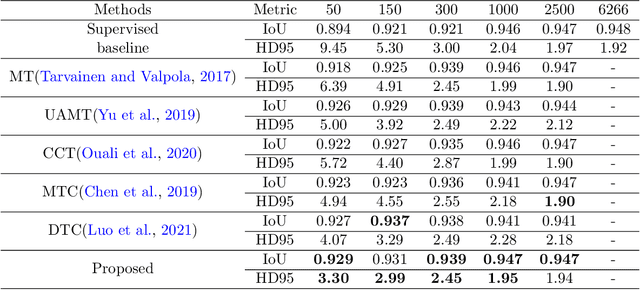
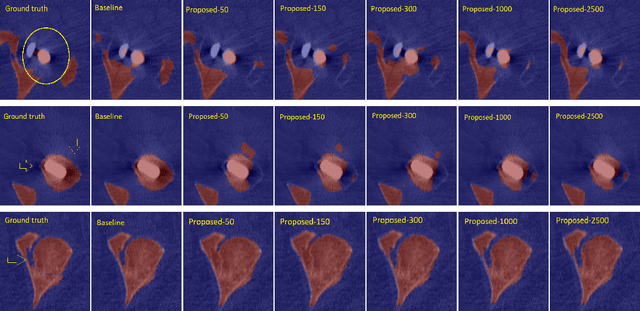

Abstract:Deep convolutional neural networks are widely used in medical image segmentation but require many labeled images for training. Annotating three-dimensional medical images is a time-consuming and costly process. To overcome this limitation, we propose a novel semi-supervised segmentation method that leverages mostly unlabeled images and a small set of labeled images in training. Our approach involves assessing prediction uncertainty to identify reliable predictions on unlabeled voxels from the teacher model. These voxels serve as pseudo-labels for training the student model. In voxels where the teacher model produces unreliable predictions, pseudo-labeling is carried out based on voxel-wise embedding correspondence using reference voxels from labeled images. We applied this method to automate hip bone segmentation in CT images, achieving notable results with just 4 CT scans. The proposed approach yielded a Hausdorff distance with 95th percentile (HD95) of 3.30 and IoU of 0.929, surpassing existing methods achieving HD95 (4.07) and IoU (0.927) at their best.
Emphysema Subtyping on Thoracic Computed Tomography Scans using Deep Neural Networks
Sep 05, 2023Abstract:Accurate identification of emphysema subtypes and severity is crucial for effective management of COPD and the study of disease heterogeneity. Manual analysis of emphysema subtypes and severity is laborious and subjective. To address this challenge, we present a deep learning-based approach for automating the Fleischner Society's visual score system for emphysema subtyping and severity analysis. We trained and evaluated our algorithm using 9650 subjects from the COPDGene study. Our algorithm achieved the predictive accuracy at 52\%, outperforming a previously published method's accuracy of 45\%. In addition, the agreement between the predicted scores of our method and the visual scores was good, where the previous method obtained only moderate agreement. Our approach employs a regression training strategy to generate categorical labels while simultaneously producing high-resolution localized activation maps for visualizing the network predictions. By leveraging these dense activation maps, our method possesses the capability to compute the percentage of emphysema involvement per lung in addition to categorical severity scores. Furthermore, the proposed method extends its predictive capabilities beyond centrilobular emphysema to include paraseptal emphysema subtypes.
Structure and position-aware graph neural network for airway labeling
Jan 12, 2022



Abstract:We present a novel graph-based approach for labeling the anatomical branches of a given airway tree segmentation. The proposed method formulates airway labeling as a branch classification problem in the airway tree graph, where branch features are extracted using convolutional neural networks (CNN) and enriched using graph neural networks. Our graph neural network is structure-aware by having each node aggregate information from its local neighbors and position-aware by encoding node positions in the graph. We evaluated the proposed method on 220 airway trees from subjects with various severity stages of Chronic Obstructive Pulmonary Disease (COPD). The results demonstrate that our approach is computationally efficient and significantly improves branch classification performance than the baseline method. The overall average accuracy of our method reaches 91.18\% for labeling all 18 segmental airway branches, compared to 83.83\% obtained by the standard CNN method. We published our source code at https://github.com/DIAGNijmegen/spgnn. The proposed algorithm is also publicly available at https://grand-challenge.org/algorithms/airway-anatomical-labeling/.
Deep Clustering Activation Maps for Emphysema Subtyping
Jun 01, 2021

Abstract:We propose a deep learning clustering method that exploits dense features from a segmentation network for emphysema subtyping from computed tomography (CT) scans. Using dense features enables high-resolution visualization of image regions corresponding to the cluster assignment via dense clustering activation maps (dCAMs). This approach provides model interpretability. We evaluated clustering results on 500 subjects from the COPDGenestudy, where radiologists manually annotated emphysema sub-types according to their visual CT assessment. We achieved a 43% unsupervised clustering accuracy, outperforming our baseline at 41% and yielding results comparable to supervised classification at 45%. The proposed method also offers a better cluster formation than the baseline, achieving0.54 in silhouette coefficient and 0.55 in David-Bouldin scores.
Dense Regression Activation Maps For Lesion Segmentation in CT scans of COVID-19 patients
May 25, 2021



Abstract:Automatic lesion segmentation on thoracic CT enables rapid quantitative analysis of lung involvement in COVID- 19 infections. Obtaining voxel-level annotations for training segmentation networks is prohibitively expensive. Therefore we propose a weakly-supervised segmentation method based on dense regression activation maps (dRAM). Most advanced weakly supervised segmentation approaches exploit class activation maps (CAMs) to localize objects generated from high-level semantic features at a coarse resolution. As a result, CAMs provide coarse outlines that do not align precisely with the object segmentations. Instead, we exploit dense features from a segmentation network to compute dense regression activation maps (dRAMs) for preserving local details. During training, dRAMs are pooled lobe-wise to regress the per-lobe lesion percentage. In such a way, the network achieves additional information regarding the lesion quantification in comparison with the classification approach. Furthermore, we refine dRAMs based on an attention module and dense conditional random field trained together with the main regression task. The refined dRAMs are served as the pseudo labels for training a final segmentation network. When evaluated on 69 CT scans, our method substantially improves the intersection over union from 0.335 in the CAM-based weakly supervised segmentation method to 0.495.
Relational Modeling for Robust and Efficient Pulmonary Lobe Segmentation in CT Scans
May 12, 2020



Abstract:Pulmonary lobe segmentation in computed tomography scans is essential for regional assessment of pulmonary diseases. Recent works based on convolution neural networks have achieved good performance for this task. However, they are still limited in capturing structured relationships due to the nature of convolution. The shape of the pulmonary lobes affect each other and their borders relate to the appearance of other structures, such as vessels, airways, and the pleural wall. We argue that such structural relationships play a critical role in the accurate delineation of pulmonary lobes when the lungs are affected by diseases such as COVID-19 or COPD. In this paper, we propose a relational approach (RTSU-Net) that leverages structured relationships by introducing a novel non-local neural network module. The proposed module learns both visual and geometric relationships among all convolution features to produce self-attention weights. With a limited amount of training data available from COVID-19 subjects, we initially train and validate RTSU-Net on a cohort of 5000 subjects from the COPDGene study (4000 for training and 1000 for evaluation). Using models pre-trained on COPDGene, we apply transfer learning to retrain and evaluate RTSU-Net on 470 COVID-19 suspects (370 for retraining and 100 for evaluation). Experimental results show that RTSU-Net outperforms three baselines and performs robustly on cases with severe lung infection due to COVID-19.
2.75D Convolutional Neural Network for Pulmonary Nodule Classification in Chest CT
Feb 11, 2020

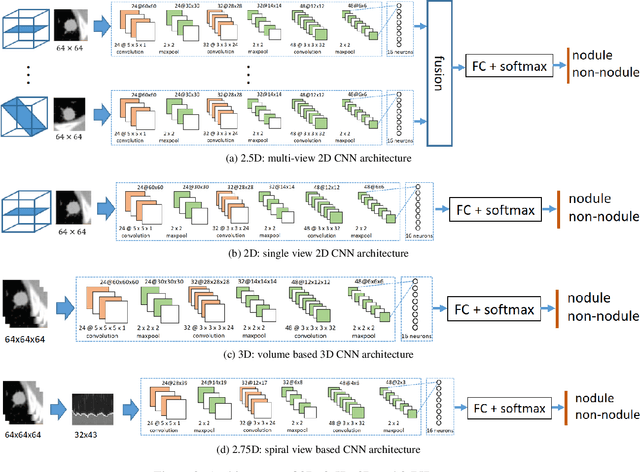

Abstract:Early detection and classification of pulmonary nodules in Chest Computed tomography (CT) images is an essential step for effective treatment of lung cancer. However, due to the large volume of CT data, finding nodules in chest CT is a time consuming thus error prone task for radiologists. Benefited from the recent advances in Convolutional Neural Networks (ConvNets), many algorithms based on ConvNets for automatic nodule detection have been proposed. According to the data representation in their input, these algorithms can be further categorized into: 2D, 3D and 2.5D which uses a combination of 2D images to approximate 3D information. Leveraging 3D spatial and contextual information, the method using 3D input generally outperform that based on 2D or 2.5D input, whereas its large memory footprints becomes the bottleneck for many applications. In this paper, we propose a novel 2D data representation of a 3D CT volume, which is constructed by spiral scanning a set of radials originated from the 3D volume center, referred to as the 2.75D. Comparing to the 2.5D, the 2.75D representation captures omni-directional spatial information of a 3D volume. Based on 2.75D representation of 3D nodule candidates in Chest CT, we train a convolutional neural network to perform the false positive reduction in the nodule detection pipeline. We evaluate the nodule false positive reduction system on the LUNA16 data set which contains 1186 nodules out of 551,065 candidates. By comparing 2.75D with 2D, 2.5D and 3D, we show that our system using 2.75D input outperforms 2D and 2.5D, yet slightly inferior to the systems using 3D input. The proposed strategy dramatically reduces the memory consumption thus allow fast inference and training by enabling larger number of batches comparing to the methods using 3D input.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge