Luuk H. Boulogne
The STOIC2021 COVID-19 AI challenge: applying reusable training methodologies to private data
Jun 25, 2023Abstract:Challenges drive the state-of-the-art of automated medical image analysis. The quantity of public training data that they provide can limit the performance of their solutions. Public access to the training methodology for these solutions remains absent. This study implements the Type Three (T3) challenge format, which allows for training solutions on private data and guarantees reusable training methodologies. With T3, challenge organizers train a codebase provided by the participants on sequestered training data. T3 was implemented in the STOIC2021 challenge, with the goal of predicting from a computed tomography (CT) scan whether subjects had a severe COVID-19 infection, defined as intubation or death within one month. STOIC2021 consisted of a Qualification phase, where participants developed challenge solutions using 2000 publicly available CT scans, and a Final phase, where participants submitted their training methodologies with which solutions were trained on CT scans of 9724 subjects. The organizers successfully trained six of the eight Final phase submissions. The submitted codebases for training and running inference were released publicly. The winning solution obtained an area under the receiver operating characteristic curve for discerning between severe and non-severe COVID-19 of 0.815. The Final phase solutions of all finalists improved upon their Qualification phase solutions.HSUXJM-TNZF9CHSUXJM-TNZF9C
Automated Estimation of Total Lung Volume using Chest Radiographs and Deep Learning
May 03, 2021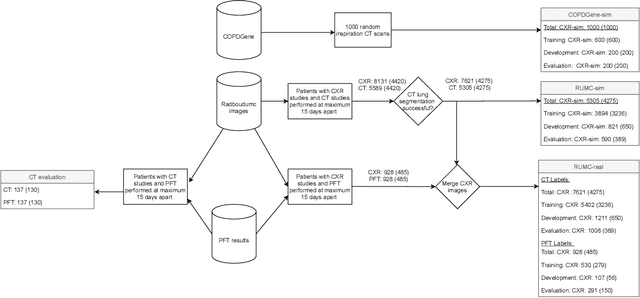
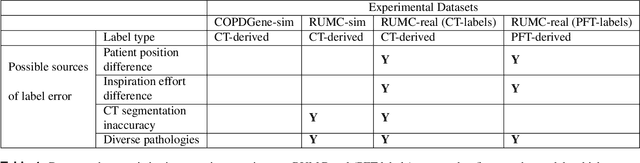
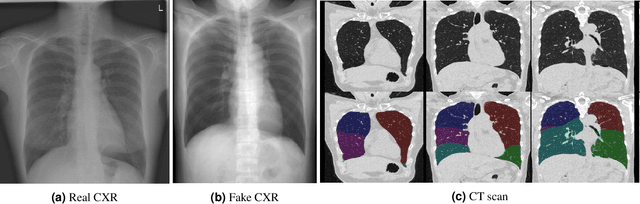
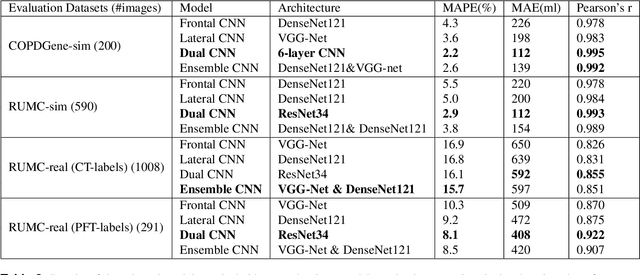
Abstract:Total lung volume is an important quantitative biomarker and is used for the assessment of restrictive lung diseases. In this study, we investigate the performance of several deep-learning approaches for automated measurement of total lung volume from chest radiographs. 7621 posteroanterior and lateral view chest radiographs (CXR) were collected from patients with chest CT available. Similarly, 928 CXR studies were chosen from patients with pulmonary function test (PFT) results. The reference total lung volume was calculated from lung segmentation on CT or PFT data, respectively. This dataset was used to train deep-learning architectures to predict total lung volume from chest radiographs. The experiments were constructed in a step-wise fashion with increasing complexity to demonstrate the effect of training with CT-derived labels only and the sources of error. The optimal models were tested on 291 CXR studies with reference lung volume obtained from PFT. The optimal deep-learning regression model showed an MAE of 408 ml and a MAPE of 8.1\% and Pearson's r = 0.92 using both frontal and lateral chest radiographs as input. CT-derived labels were useful for pre-training but the optimal performance was obtained by fine-tuning the network with PFT-derived labels. We demonstrate, for the first time, that state-of-the-art deep learning solutions can accurately measure total lung volume from plain chest radiographs. The proposed model can be used to obtain total lung volume from routinely acquired chest radiographs at no additional cost and could be a useful tool to identify trends over time in patients referred regularly for chest x-rays.
Improving Automated COVID-19 Grading with Convolutional Neural Networks in Computed Tomography Scans: An Ablation Study
Sep 21, 2020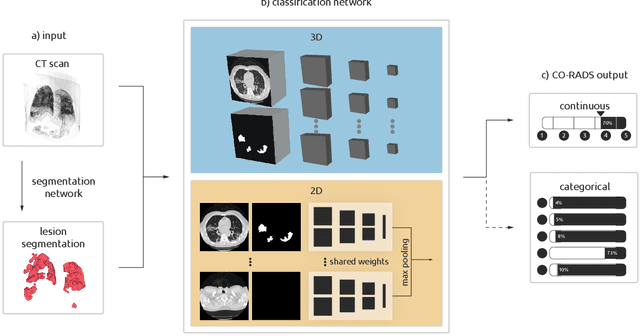
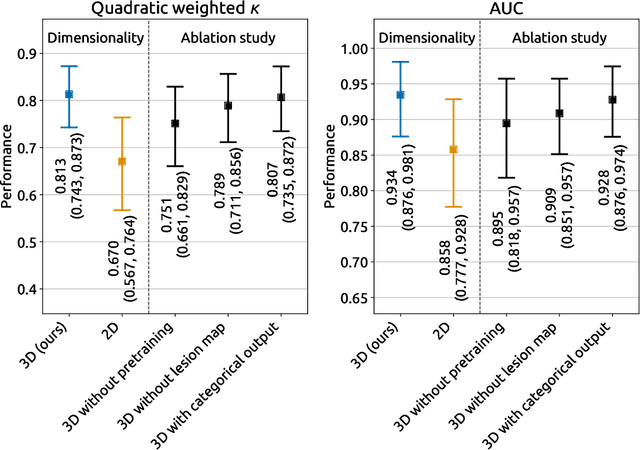
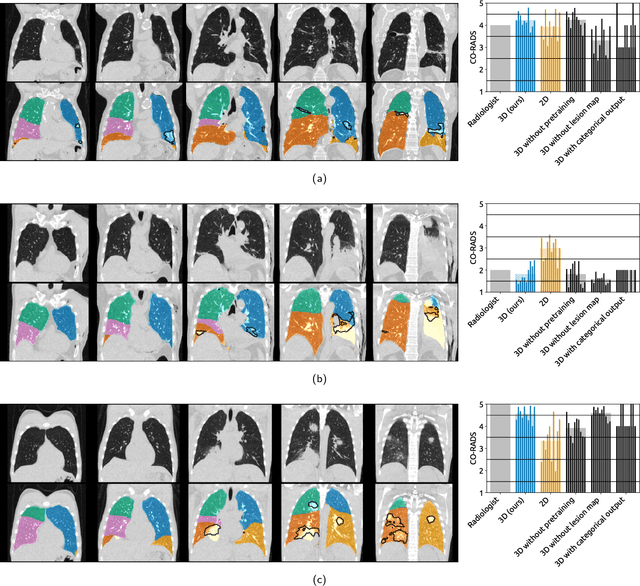
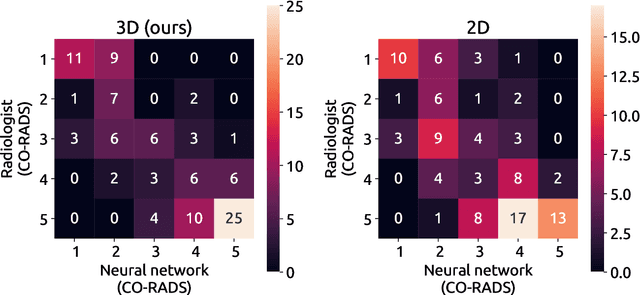
Abstract:Amidst the ongoing pandemic, several studies have shown that COVID-19 classification and grading using computed tomography (CT) images can be automated with convolutional neural networks (CNNs). Many of these studies focused on reporting initial results of algorithms that were assembled from commonly used components. The choice of these components was often pragmatic rather than systematic. For instance, several studies used 2D CNNs even though these might not be optimal for handling 3D CT volumes. This paper identifies a variety of components that increase the performance of CNN-based algorithms for COVID-19 grading from CT images. We investigated the effectiveness of using a 3D CNN instead of a 2D CNN, of using transfer learning to initialize the network, of providing automatically computed lesion maps as additional network input, and of predicting a continuous instead of a categorical output. A 3D CNN with these components achieved an area under the ROC curve (AUC) of 0.934 on our test set of 105 CT scans and an AUC of 0.923 on a publicly available set of 742 CT scans, a substantial improvement in comparison with a previously published 2D CNN. An ablation study demonstrated that in addition to using a 3D CNN instead of a 2D CNN transfer learning contributed the most and continuous output contributed the least to improving the model performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge