Matthieu Rutten
Lumbar spine segmentation in MR images: a dataset and a public benchmark
Jun 22, 2023Abstract:This paper presents a large publicly available multi-center lumbar spine magnetic resonance imaging (MRI) dataset with reference segmentations of vertebrae, intervertebral discs (IVDs), and spinal canal. The dataset includes 447 sagittal T1 and T2 MRI series from 218 patients with a history of low back pain. It was collected from four different hospitals and was divided into a training (179 patients) and validation (39 patients) set. An iterative data annotation approach was used by training a segmentation algorithm on a small part of the dataset, enabling semi-automatic segmentation of the remaining images. The algorithm provided an initial segmentation, which was subsequently reviewed, manually corrected, and added to the training data. We provide reference performance values for this baseline algorithm and nnU-Net, which performed comparably. We set up a continuous segmentation challenge to allow for a fair comparison of different segmentation algorithms. This study may encourage wider collaboration in the field of spine segmentation, and improve the diagnostic value of lumbar spine MRI.
Deep Learning with robustness to missing data: A novel approach to the detection of COVID-19
Mar 25, 2021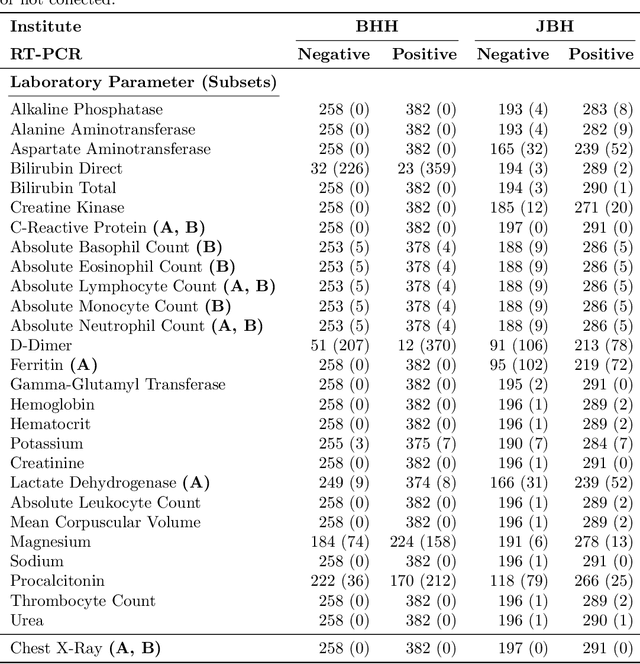
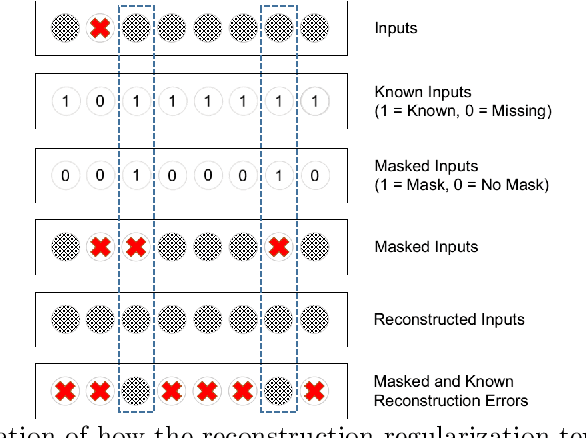
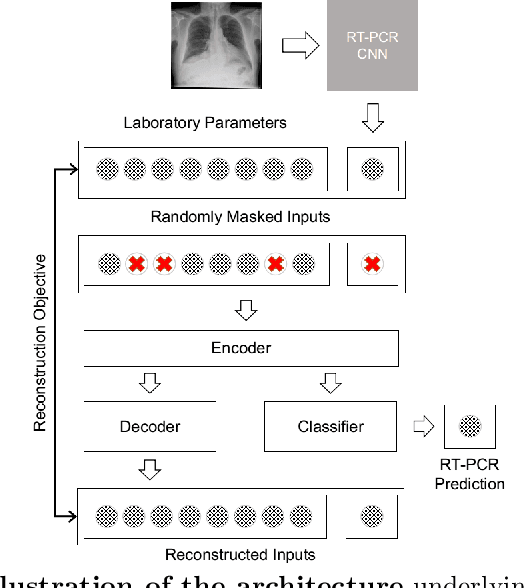
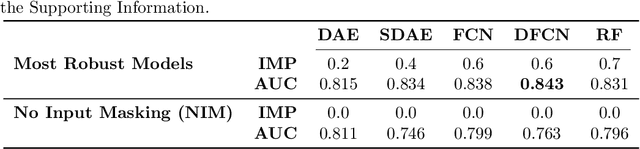
Abstract:In the context of the current global pandemic and the limitations of the RT-PCR test, we propose a novel deep learning architecture, DFCN, (Denoising Fully Connected Network) for the detection of COVID-19 using laboratory tests and chest x-rays. Since medical facilities around the world differ enormously in what laboratory tests or chest imaging may be available, DFCN is designed to be robust to missing input data. An ablation study extensively evaluates the performance benefits of the DFCN architecture as well as its robustness to missing inputs. Data from 1088 patients with confirmed RT-PCR results are obtained from two independent medical facilities. The data collected includes results from 27 laboratory tests and a chest x-ray scored by a deep learning network. Training and test datasets are defined based on the source medical facility. Data is made publicly available. The performance of DFCN in predicting the RT-PCR result is compared with 3 related architectures as well as a Random Forest baseline. All models are trained with varying levels of masked input data to encourage robustness to missing inputs. Missing data is simulated at test time by masking inputs randomly. Using area under the receiver operating curve (AUC) as a metric, DFCN outperforms all other models with statistical significance using random subsets of input data with 2-27 available inputs. When all 28 inputs are available DFCN obtains an AUC of 0.924, higher than achieved by any other model. Furthermore, with clinically meaningful subsets of parameters consisting of just 6 and 7 inputs respectively, DFCN also achieves higher AUCs than any other model, with values of 0.909 and 0.919.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge