Bob D. de Vos
Generative Models for Reproducible Coronary Calcium Scoring
May 24, 2022



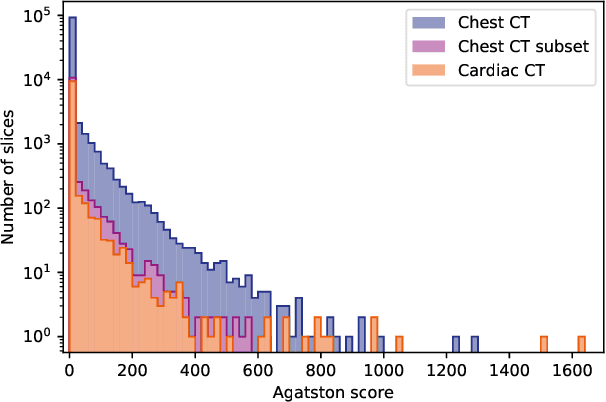
Abstract:Purpose: Coronary artery calcium (CAC) score, i.e. the amount of CAC quantified in CT, is a strong and independent predictor of coronary heart disease (CHD) events. However, CAC scoring suffers from limited interscan reproducibility, which is mainly due to the clinical definition requiring application of a fixed intensity level threshold for segmentation of calcifications. This limitation is especially pronounced in non-ECG-synchronized CT where lesions are more impacted by cardiac motion and partial volume effects. Therefore, we propose a CAC quantification method that does not require a threshold for segmentation of CAC. Approach: Our method utilizes a generative adversarial network where a CT with CAC is decomposed into an image without CAC and an image showing only CAC. The method, using a CycleGAN, was trained using 626 low-dose chest CTs and 514 radiotherapy treatment planning CTs. Interscan reproducibility was compared to clinical calcium scoring in radiotherapy treatment planning CTs of 1,662 patients, each having two scans. Results: A lower relative interscan difference in CAC mass was achieved by the proposed method: 47% compared to 89% manual clinical calcium scoring. The intraclass correlation coefficient of Agatston scores was 0.96 for the proposed method compared to 0.91 for automatic clinical calcium scoring. Conclusions: The increased interscan reproducibility achieved by our method may lead to increased reliability of CHD risk categorization and improved accuracy of CHD event prediction.
* In press
Autoencoding Low-Resolution MRI for Semantically Smooth Interpolation of Anisotropic MRI
Feb 18, 2022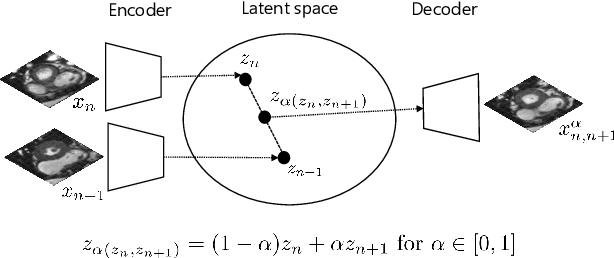
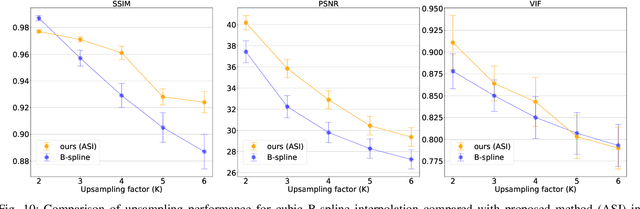
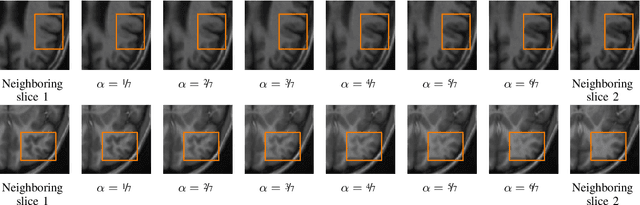
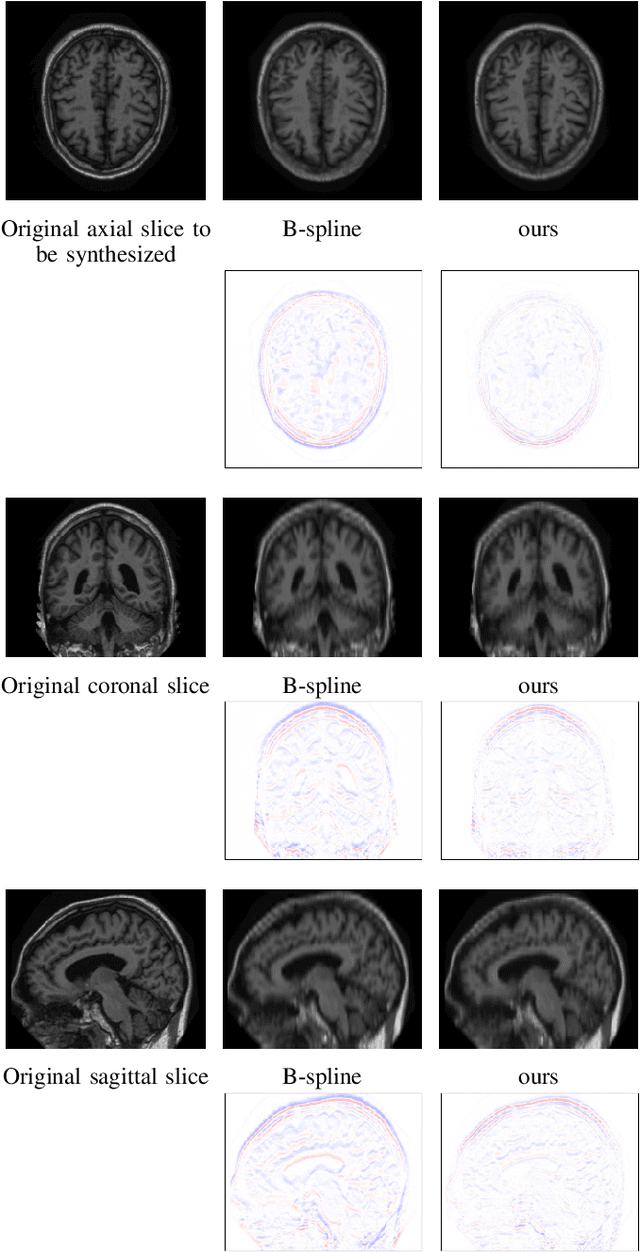
Abstract:High-resolution medical images are beneficial for analysis but their acquisition may not always be feasible. Alternatively, high-resolution images can be created from low-resolution acquisitions using conventional upsampling methods, but such methods cannot exploit high-level contextual information contained in the images. Recently, better performing deep-learning based super-resolution methods have been introduced. However, these methods are limited by their supervised character, i.e. they require high-resolution examples for training. Instead, we propose an unsupervised deep learning semantic interpolation approach that synthesizes new intermediate slices from encoded low-resolution examples. To achieve semantically smooth interpolation in through-plane direction, the method exploits the latent space generated by autoencoders. To generate new intermediate slices, latent space encodings of two spatially adjacent slices are combined using their convex combination. Subsequently, the combined encoding is decoded to an intermediate slice. To constrain the model, a notion of semantic similarity is defined for a given dataset. For this, a new loss is introduced that exploits the spatial relationship between slices of the same volume. During training, an existing in-between slice is generated using a convex combination of its neighboring slice encodings. The method was trained and evaluated using publicly available cardiac cine, neonatal brain and adult brain MRI scans. In all evaluations, the new method produces significantly better results in terms of Structural Similarity Index Measure and Peak Signal-to-Noise Ratio (p< 0.001 using one-sided Wilcoxon signed-rank test) than a cubic B-spline interpolation approach. Given the unsupervised nature of the method, high-resolution training data is not required and hence, the method can be readily applied in clinical settings.
AI for Calcium Scoring
May 24, 2021



Abstract:Calcium scoring, a process in which arterial calcifications are detected and quantified in CT, is valuable in estimating the risk of cardiovascular disease events. Especially when used to quantify the extent of calcification in the coronary arteries, it is a strong and independent predictor of coronary heart disease events. Advances in artificial intelligence (AI)-based image analysis have produced a multitude of automatic calcium scoring methods. While most early methods closely follow standard calcium scoring accepted in clinic, recent approaches extend this procedure to enable faster or more reproducible calcium scoring. This chapter provides an introduction to AI for calcium scoring, and an overview of the developed methods and their applications. We conclude with a discussion on AI methods in calcium scoring and propose potential directions for future research.
Automatic segmentation with detection of local segmentation failures in cardiac MRI
Nov 13, 2020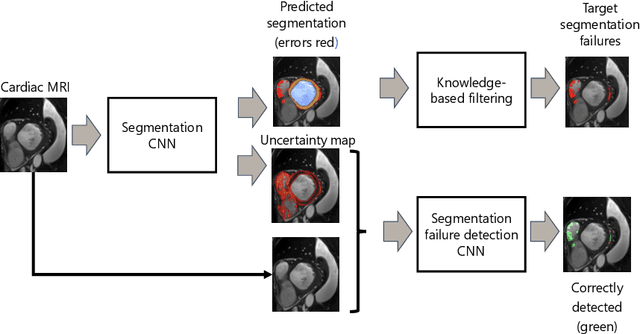
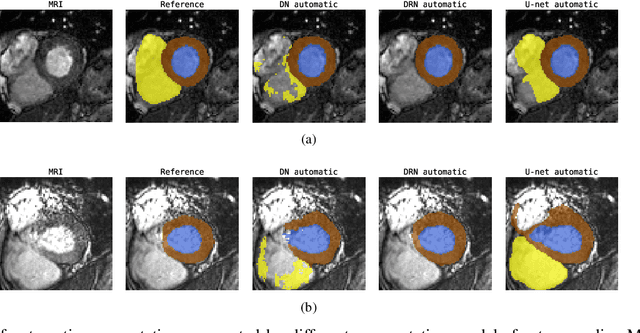
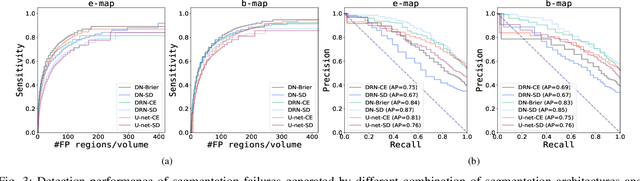
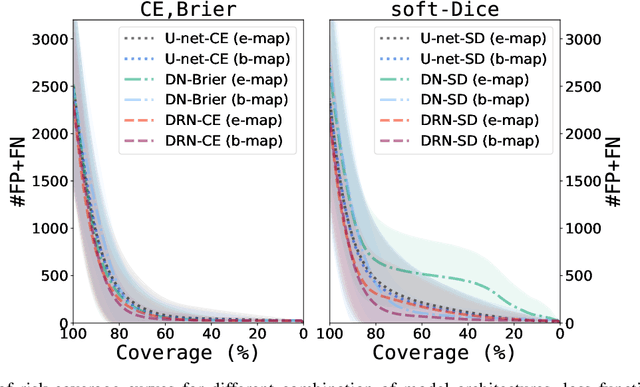
Abstract:Segmentation of cardiac anatomical structures in cardiac magnetic resonance images (CMRI) is a prerequisite for automatic diagnosis and prognosis of cardiovascular diseases. To increase robustness and performance of segmentation methods this study combines automatic segmentation and assessment of segmentation uncertainty in CMRI to detect image regions containing local segmentation failures. Three state-of-the-art convolutional neural networks (CNN) were trained to automatically segment cardiac anatomical structures and obtain two measures of predictive uncertainty: entropy and a measure derived by MC-dropout. Thereafter, using the uncertainties another CNN was trained to detect local segmentation failures that potentially need correction by an expert. Finally, manual correction of the detected regions was simulated. Using publicly available CMR scans from the MICCAI 2017 ACDC challenge, the impact of CNN architecture and loss function for segmentation, and the uncertainty measure was investigated. Performance was evaluated using the Dice coefficient and 3D Hausdorff distance between manual and automatic segmentation. The experiments reveal that combining automatic segmentation with simulated manual correction of detected segmentation failures leads to statistically significant performance increase.
Unsupervised Super-Resolution: Creating High-Resolution Medical Images from Low-Resolution Anisotropic Examples
Oct 25, 2020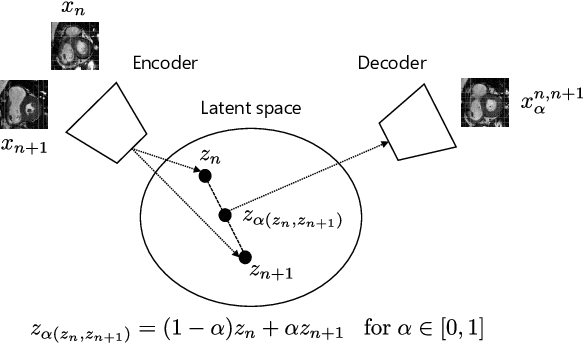
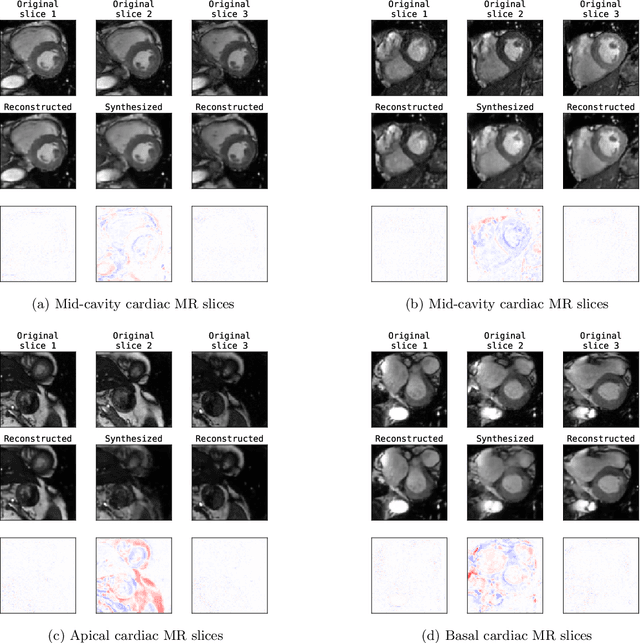
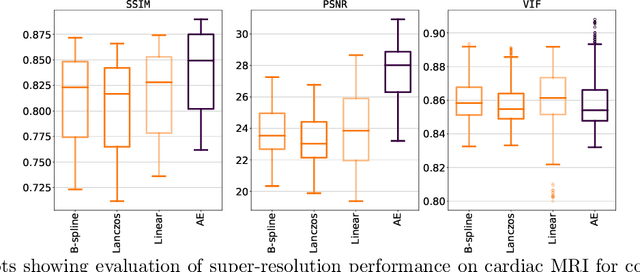
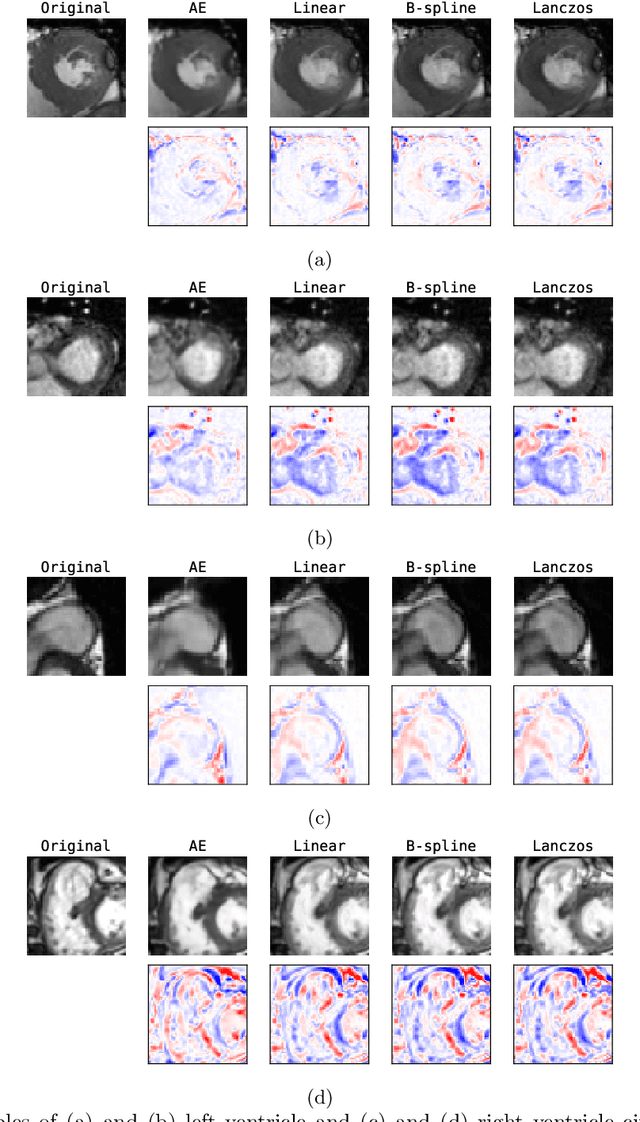
Abstract:Although high resolution isotropic 3D medical images are desired in clinical practice, their acquisition is not always feasible. Instead, lower resolution images are upsampled to higher resolution using conventional interpolation methods. Sophisticated learning-based super-resolution approaches are frequently unavailable in clinical setting, because such methods require training with high-resolution isotropic examples. To address this issue, we propose a learning-based super-resolution approach that can be trained using solely anisotropic images, i.e. without high-resolution ground truth data. The method exploits the latent space, generated by autoencoders trained on anisotropic images, to increase spatial resolution in low-resolution images. The method was trained and evaluated using 100 publicly available cardiac cine MR scans from the Automated Cardiac Diagnosis Challenge (ACDC). The quantitative results show that the proposed method performs better than conventional interpolation methods. Furthermore, the qualitative results indicate that especially finer cardiac structures are synthesized with high quality. The method has the potential to be applied to other anatomies and modalities and can be easily applied to any 3D anisotropic medical image dataset.
Deep Group-wise Variational Diffeomorphic Image Registration
Oct 01, 2020

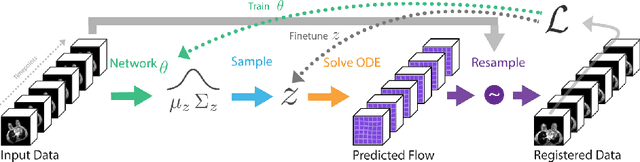
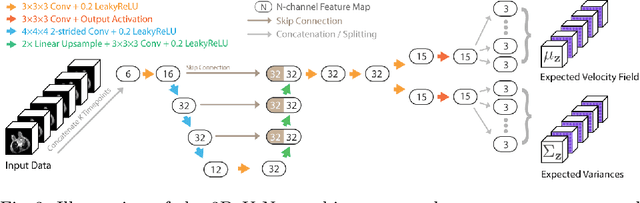
Abstract:Deep neural networks are increasingly used for pair-wise image registration. We propose to extend current learning-based image registration to allow simultaneous registration of multiple images. To achieve this, we build upon the pair-wise variational and diffeomorphic VoxelMorph approach and present a general mathematical framework that enables both registration of multiple images to their geodesic average and registration in which any of the available images can be used as a fixed image. In addition, we provide a likelihood based on normalized mutual information, a well-known image similarity metric in registration, between multiple images, and a prior that allows for explicit control over the viscous fluid energy to effectively regularize deformations. We trained and evaluated our approach using intra-patient registration of breast MRI and Thoracic 4DCT exams acquired over multiple time points. Comparison with Elastix and VoxelMorph demonstrates competitive quantitative performance of the proposed method in terms of image similarity and reference landmark distances at significantly faster registration.
Deep Learning-Based Regression and Classification for Automatic Landmark Localization in Medical Images
Jul 10, 2020



Abstract:In this study, we propose a fast and accurate method to automatically localize anatomical landmarks in medical images. We employ a global-to-local localization approach using fully convolutional neural networks (FCNNs). First, a global FCNN localizes multiple landmarks through the analysis of image patches, performing regression and classification simultaneously. In regression, displacement vectors pointing from the center of image patches towards landmark locations are determined. In classification, presence of landmarks of interest in the patch is established. Global landmark locations are obtained by averaging the predicted displacement vectors, where the contribution of each displacement vector is weighted by the posterior classification probability of the patch that it is pointing from. Subsequently, for each landmark localized with global localization, local analysis is performed. Specialized FCNNs refine the global landmark locations by analyzing local sub-images in a similar manner, i.e. by performing regression and classification simultaneously and combining the results. Evaluation was performed through localization of 8 anatomical landmarks in CCTA scans, 2 landmarks in olfactory MR scans, and 19 landmarks in cephalometric X-rays. We demonstrate that the method performs similarly to a second observer and is able to localize landmarks in a diverse set of medical images, differing in image modality, image dimensionality, and anatomical coverage.
Direct Automatic Coronary Calcium Scoring in Cardiac and Chest CT
Feb 12, 2019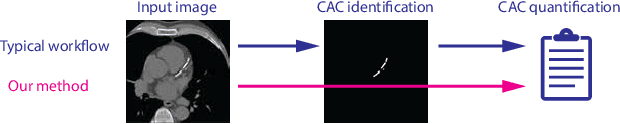
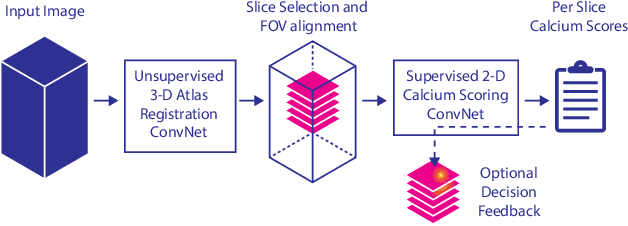
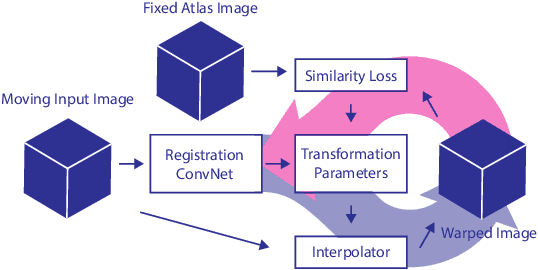
Abstract:Cardiovascular disease (CVD) is the global leading cause of death. A strong risk factor for CVD events is the amount of coronary artery calcium (CAC). To meet demands of the increasing interest in quantification of CAC, i.e. coronary calcium scoring, especially as an unrequested finding for screening and research, automatic methods have been proposed. Current automatic calcium scoring methods are relatively computationally expensive and only provide scores for one type of CT. To address this, we propose a computationally efficient method that employs two ConvNets: the first performs registration to align the fields of view of input CTs and the second performs direct regression of the calcium score, thereby circumventing time-consuming intermediate CAC segmentation. Optional decision feedback provides insight in the regions that contributed to the calcium score. Experiments were performed using 903 cardiac CT and 1,687 chest CT scans. The method predicted calcium scores in less than 0.3 s. Intra-class correlation coefficient between predicted and manual calcium scores was 0.98 for both cardiac and chest CT. The method showed almost perfect agreement between automatic and manual CVD risk categorization in both datasets, with a linearly weighted Cohen's kappa of 0.95 in cardiac CT and 0.93 in chest CT. Performance is similar to that of state-of-the-art methods, but the proposed method is hundreds of times faster. By providing visual feedback, insight is given in the decision process, making it readily implementable in clinical and research settings.
Response monitoring of breast cancer on DCE-MRI using convolutional neural network-generated seed points and constrained volume growing
Nov 22, 2018Abstract:Response of breast cancer to neoadjuvant chemotherapy (NAC) can be monitored using the change in visible tumor on magnetic resonance imaging (MRI). In our current workflow, seed points are manually placed in areas of enhancement likely to contain cancer. A constrained volume growing method uses these manually placed seed points as input and generates a tumor segmentation. This method is rigorously validated using complete pathological embedding. In this study, we propose to exploit deep learning for fast and automatic seed point detection, replacing manual seed point placement in our existing and well-validated workflow. The seed point generator was developed in early breast cancer patients with pathology-proven segmentations (N=100), operated shortly after MRI. It consisted of an ensemble of three independently trained fully convolutional dilated neural networks that classified breast voxels as tumor or non-tumor. Subsequently, local maxima were used as seed points for volume growing in patients receiving NAC (N=10). The percentage of tumor volume change was evaluated against semi-automatic segmentations. The primary cancer was localized in 95% of the tumors at the cost of 0.9 false positive per patient. False positives included focally enhancing regions of unknown origin and parts of the intramammary blood vessels. Volume growing from the seed points showed a median tumor volume decrease of 70% (interquartile range: 50%-77%), comparable to the semi-automatic segmentations (median: 70%, interquartile range 23%-76%). To conclude, a fast and automatic seed point generator was developed, fully automating a well-validated semi-automatic workflow for response monitoring of breast cancer to neoadjuvant chemotherapy.
Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT
Oct 09, 2018Abstract:Morphological analysis and identification of pathologies in the aorta are important for cardiovascular diagnosis and risk assessment in patients. Manual annotation is time-consuming and cumbersome in CT scans acquired without contrast enhancement and with low radiation dose. Hence, we propose an automatic method to segment the ascending aorta, the aortic arch and the thoracic descending aorta in low-dose chest CT without contrast enhancement. Segmentation was performed using a dilated convolutional neural network (CNN), with a receptive field of 131X131 voxels, that classified voxels in axial, coronal and sagittal image slices. To obtain a final segmentation, the obtained probabilities of the three planes were averaged per class, and voxels were subsequently assigned to the class with the highest class probability. Two-fold cross-validation experiments were performed where ten scans were used to train the network and another ten to evaluate the performance. Dice coefficients of 0.83, 0.86 and 0.88, and Average Symmetrical Surface Distances (ASSDs) of 2.44, 1.56 and 1.87 mm were obtained for the ascending aorta, the aortic arch, and the descending aorta, respectively. The results indicate that the proposed method could be used in large-scale studies analyzing the anatomical location of pathology and morphology of the thoracic aorta.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge