Bas H. M. van der Velden
for the ALFA study
BrightCookies at SemEval-2025 Task 9: Exploring Data Augmentation for Food Hazard Classification
Apr 29, 2025Abstract:This paper presents our system developed for the SemEval-2025 Task 9: The Food Hazard Detection Challenge. The shared task's objective is to evaluate explainable classification systems for classifying hazards and products in two levels of granularity from food recall incident reports. In this work, we propose text augmentation techniques as a way to improve poor performance on minority classes and compare their effect for each category on various transformer and machine learning models. We explore three word-level data augmentation techniques, namely synonym replacement, random word swapping, and contextual word insertion. The results show that transformer models tend to have a better overall performance. None of the three augmentation techniques consistently improved overall performance for classifying hazards and products. We observed a statistically significant improvement (P < 0.05) in the fine-grained categories when using the BERT model to compare the baseline with each augmented model. Compared to the baseline, the contextual words insertion augmentation improved the accuracy of predictions for the minority hazard classes by 6%. This suggests that targeted augmentation of minority classes can improve the performance of transformer models.
Federated learning in food research
Jun 10, 2024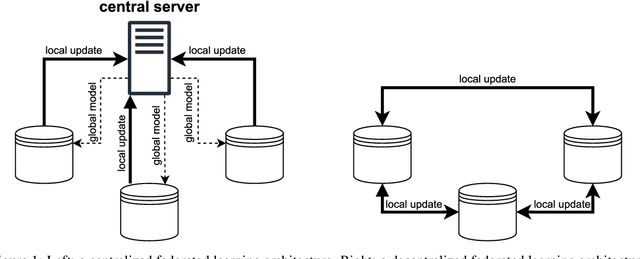
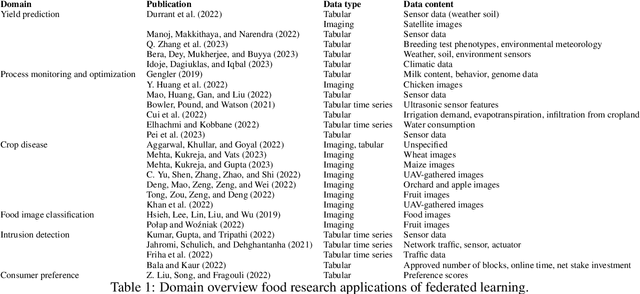
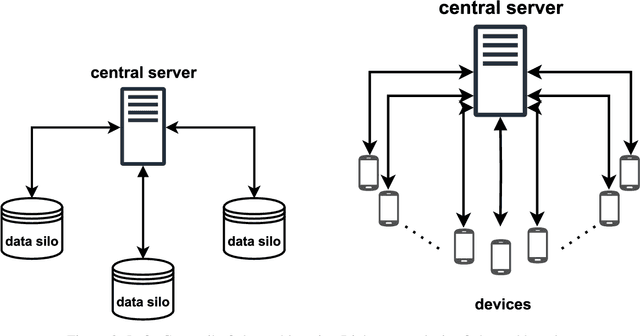

Abstract:Research in the food domain is at times limited due to data sharing obstacles, such as data ownership, privacy requirements, and regulations. While important, these obstacles can restrict data-driven methods such as machine learning. Federated learning, the approach of training models on locally kept data and only sharing the learned parameters, is a potential technique to alleviate data sharing obstacles. This systematic review investigates the use of federated learning within the food domain, structures included papers in a federated learning framework, highlights knowledge gaps, and discusses potential applications. A total of 41 papers were included in the review. The current applications include solutions to water and milk quality assessment, cybersecurity of water processing, pesticide residue risk analysis, weed detection, and fraud detection, focusing on centralized horizontal federated learning. One of the gaps found was the lack of vertical or transfer federated learning and decentralized architectures.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022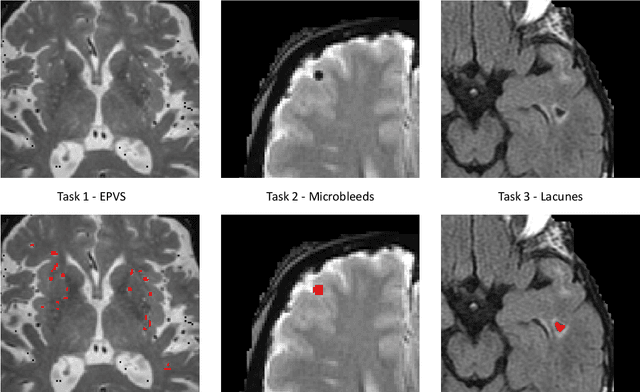
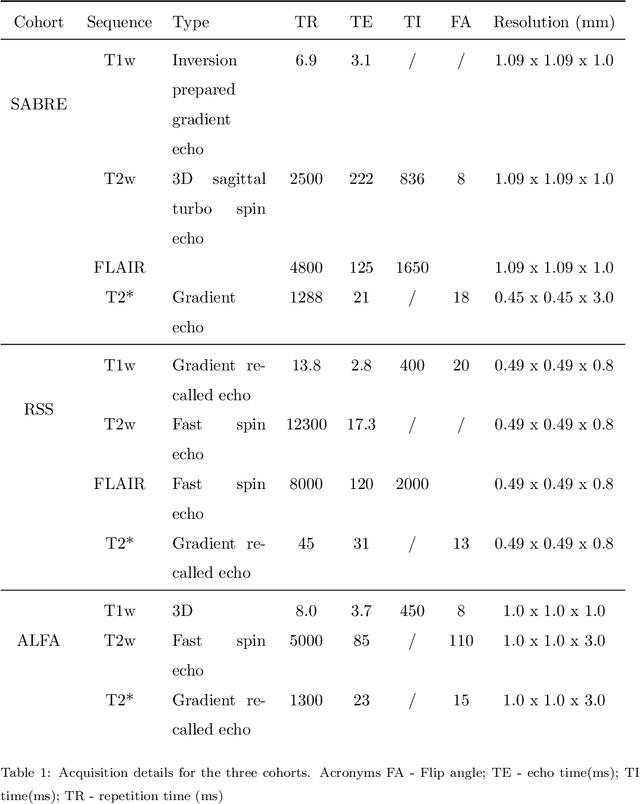
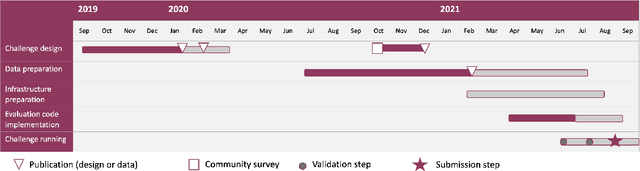
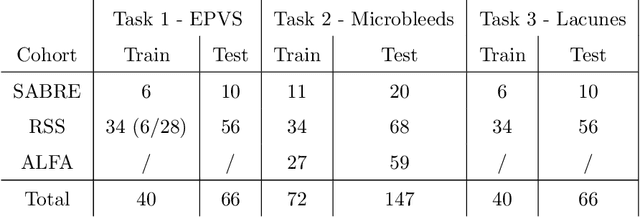
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
Multi-modal volumetric concept activation to explain detection and classification of metastatic prostate cancer on PSMA-PET/CT
Aug 04, 2022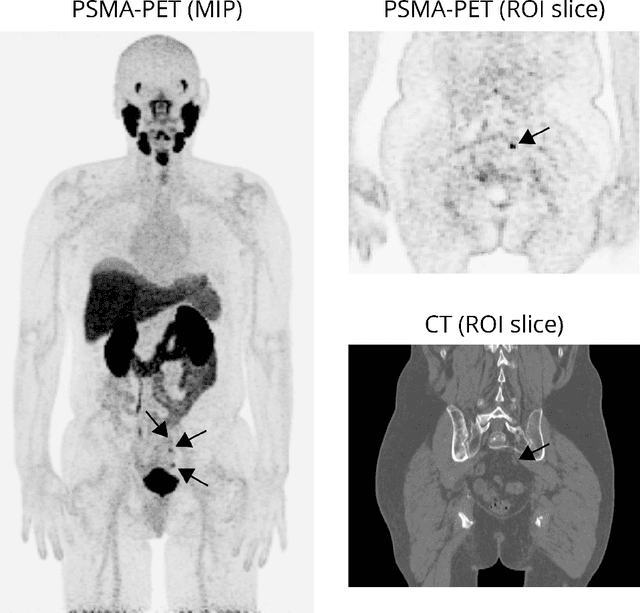

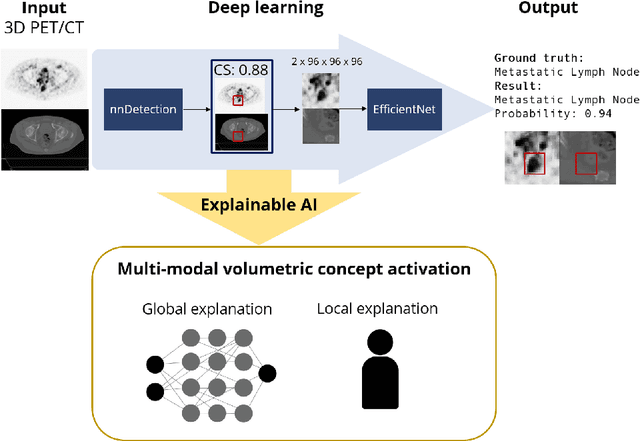
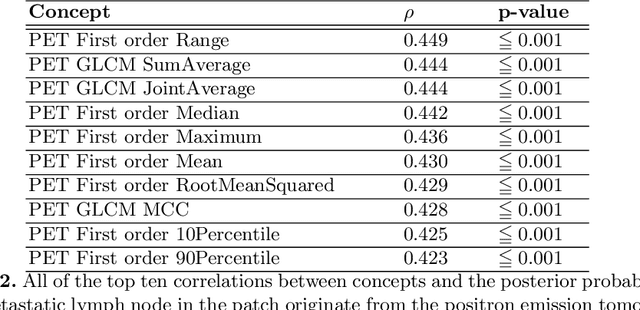
Abstract:Explainable artificial intelligence (XAI) is increasingly used to analyze the behavior of neural networks. Concept activation uses human-interpretable concepts to explain neural network behavior. This study aimed at assessing the feasibility of regression concept activation to explain detection and classification of multi-modal volumetric data. Proof-of-concept was demonstrated in metastatic prostate cancer patients imaged with positron emission tomography/computed tomography (PET/CT). Multi-modal volumetric concept activation was used to provide global and local explanations. Sensitivity was 80% at 1.78 false positive per patient. Global explanations showed that detection focused on CT for anatomical location and on PET for its confidence in the detection. Local explanations showed promise to aid in distinguishing true positives from false positives. Hence, this study demonstrated feasibility to explain detection and classification of multi-modal volumetric data using regression concept activation.
Leveraging Clinical Characteristics for Improved Deep Learning-Based Kidney Tumor Segmentation on CT
Sep 13, 2021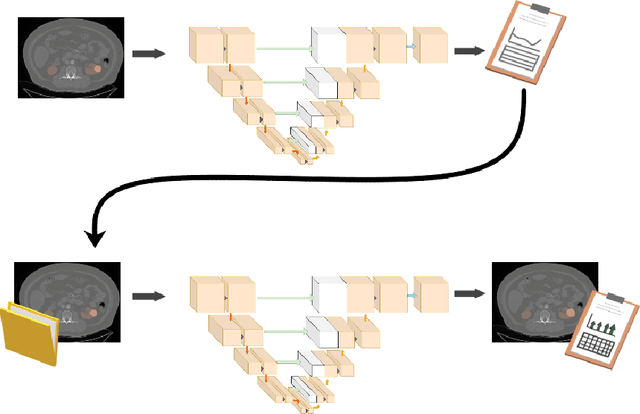

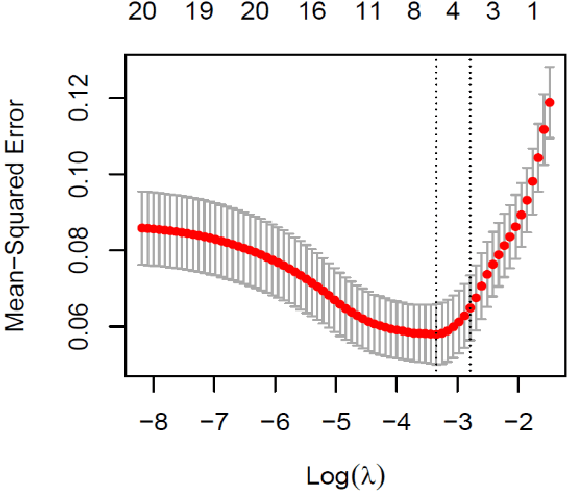

Abstract:This paper assesses whether using clinical characteristics in addition to imaging can improve automated segmentation of kidney cancer on contrast-enhanced computed tomography (CT). A total of 300 kidney cancer patients with contrast-enhanced CT scans and clinical characteristics were included. A baseline segmentation of the kidney cancer was performed using a 3D U-Net. Input to the U-Net were the contrast-enhanced CT images, output were segmentations of kidney, kidney tumors, and kidney cysts. A cognizant sampling strategy was used to leverage clinical characteristics for improved segmentation. To this end, a Least Absolute Shrinkage and Selection Operator (LASSO) was used. Segmentations were evaluated using Dice and Surface Dice. Improvement in segmentation was assessed using Wilcoxon signed rank test. The baseline 3D U-Net showed a segmentation performance of 0.90 for kidney and kidney masses, i.e., kidney, tumor, and cyst, 0.29 for kidney masses, and 0.28 for kidney tumor, while the 3D U-Net trained with cognizant sampling enhanced the segmentation performance and reached Dice scores of 0.90, 0.39, and 0.38 respectively. To conclude, the cognizant sampling strategy leveraging the clinical characteristics significantly improved kidney cancer segmentation.
MixLacune: Segmentation of lacunes of presumed vascular origin
Aug 05, 2021



Abstract:Lacunes of presumed vascular origin are fluid-filled cavities of between 3 - 15 mm in diameter, visible on T1 and FLAIR brain MRI. Quantification of lacunes relies on manual annotation or semi-automatic / interactive approaches; and almost no automatic methods exist for this task. In this work, we present a two-stage approach to segment lacunes of presumed vascular origin: (1) detection with Mask R-CNN followed by (2) segmentation with a U-Net CNN. Data originates from Task 3 of the "Where is VALDO?" challenge and consists of 40 training subjects. We report the mean DICE on the training set of 0.83 and on the validation set of 0.84. Source code is available at: https://github.com/hjkuijf/MixLacune . The docker container hjkuijf/mixlacune can be pulled from https://hub.docker.com/r/hjkuijf/mixlacune .
MixMicrobleed: Multi-stage detection and segmentation of cerebral microbleeds
Aug 05, 2021



Abstract:Cerebral microbleeds are small, dark, round lesions that can be visualised on T2*-weighted MRI or other sequences sensitive to susceptibility effects. In this work, we propose a multi-stage approach to both microbleed detection and segmentation. First, possible microbleed locations are detected with a Mask R-CNN technique. Second, at each possible microbleed location, a simple U-Net performs the final segmentation. This work used the 72 subjects as training data provided by the "Where is VALDO?" challenge of MICCAI 2021.
Explainable artificial intelligence (XAI) in deep learning-based medical image analysis
Jul 22, 2021



Abstract:With an increase in deep learning-based methods, the call for explainability of such methods grows, especially in high-stakes decision making areas such as medical image analysis. This survey presents an overview of eXplainable Artificial Intelligence (XAI) used in deep learning-based medical image analysis. A framework of XAI criteria is introduced to classify deep learning-based medical image analysis methods. Papers on XAI techniques in medical image analysis are then surveyed and categorized according to the framework and according to anatomical location. The paper concludes with an outlook of future opportunities for XAI in medical image analysis.
Interpretable deep learning regression for breast density estimation on MRI
Dec 08, 2020Abstract:Breast density, which is the ratio between fibroglandular tissue (FGT) and total breast volume, can be assessed qualitatively by radiologists and quantitatively by computer algorithms. These algorithms often rely on segmentation of breast and FGT volume. In this study, we propose a method to directly assess breast density on MRI, and provide interpretations of these assessments. We assessed breast density in 506 patients with breast cancer using a regression convolutional neural network (CNN). The input for the CNN were slices of breast MRI of 128 x 128 voxels, and the output was a continuous density value between 0 (fatty breast) and 1 (dense breast). We used 350 patients to train the CNN, 75 for validation, and 81 for independent testing. We investigated why the CNN came to its predicted density using Deep SHapley Additive exPlanations (SHAP). The density predicted by the CNN on the testing set was significantly correlated with the ground truth densities (N = 81 patients, Spearman's rho = 0.86, P < 0.001). When inspecting what the CNN based its predictions on, we found that voxels in FGT commonly had positive SHAP-values, voxels in fatty tissue commonly had negative SHAP-values, and voxels in non-breast tissue commonly had SHAP-values near zero. This means that the prediction of density is based on the structures we expect it to be based on, namely FGT and fatty tissue. To conclude, we presented an interpretable deep learning regression method for breast density estimation on MRI with promising results.
Response monitoring of breast cancer on DCE-MRI using convolutional neural network-generated seed points and constrained volume growing
Nov 22, 2018Abstract:Response of breast cancer to neoadjuvant chemotherapy (NAC) can be monitored using the change in visible tumor on magnetic resonance imaging (MRI). In our current workflow, seed points are manually placed in areas of enhancement likely to contain cancer. A constrained volume growing method uses these manually placed seed points as input and generates a tumor segmentation. This method is rigorously validated using complete pathological embedding. In this study, we propose to exploit deep learning for fast and automatic seed point detection, replacing manual seed point placement in our existing and well-validated workflow. The seed point generator was developed in early breast cancer patients with pathology-proven segmentations (N=100), operated shortly after MRI. It consisted of an ensemble of three independently trained fully convolutional dilated neural networks that classified breast voxels as tumor or non-tumor. Subsequently, local maxima were used as seed points for volume growing in patients receiving NAC (N=10). The percentage of tumor volume change was evaluated against semi-automatic segmentations. The primary cancer was localized in 95% of the tumors at the cost of 0.9 false positive per patient. False positives included focally enhancing regions of unknown origin and parts of the intramammary blood vessels. Volume growing from the seed points showed a median tumor volume decrease of 70% (interquartile range: 50%-77%), comparable to the semi-automatic segmentations (median: 70%, interquartile range 23%-76%). To conclude, a fast and automatic seed point generator was developed, fully automating a well-validated semi-automatic workflow for response monitoring of breast cancer to neoadjuvant chemotherapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge