Denis Kutnar
for the ALFA study
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022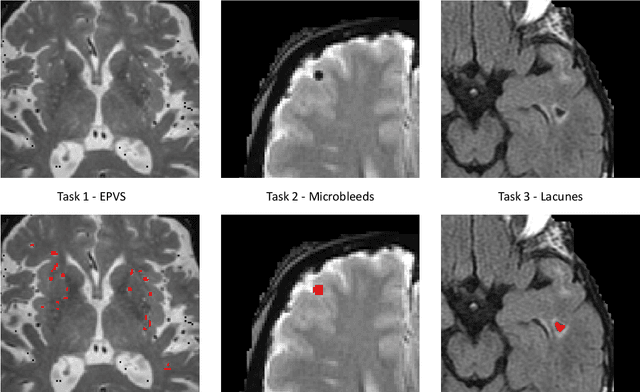
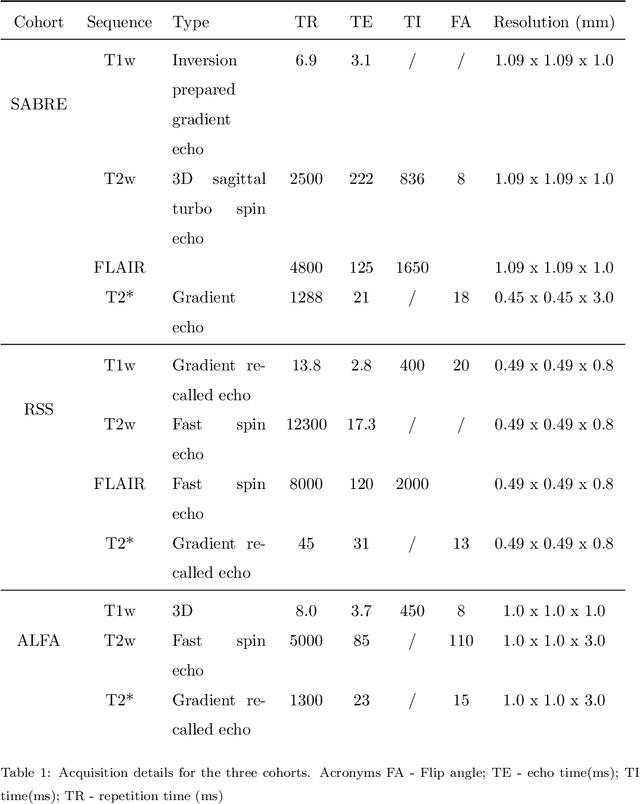
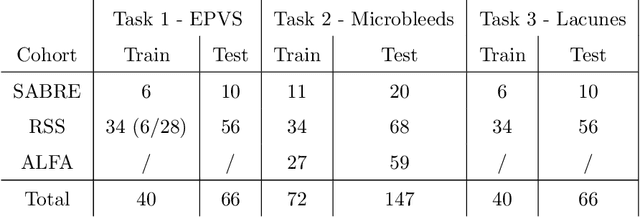
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
MixLacune: Segmentation of lacunes of presumed vascular origin
Aug 05, 2021



Abstract:Lacunes of presumed vascular origin are fluid-filled cavities of between 3 - 15 mm in diameter, visible on T1 and FLAIR brain MRI. Quantification of lacunes relies on manual annotation or semi-automatic / interactive approaches; and almost no automatic methods exist for this task. In this work, we present a two-stage approach to segment lacunes of presumed vascular origin: (1) detection with Mask R-CNN followed by (2) segmentation with a U-Net CNN. Data originates from Task 3 of the "Where is VALDO?" challenge and consists of 40 training subjects. We report the mean DICE on the training set of 0.83 and on the validation set of 0.84. Source code is available at: https://github.com/hjkuijf/MixLacune . The docker container hjkuijf/mixlacune can be pulled from https://hub.docker.com/r/hjkuijf/mixlacune .
MixMicrobleed: Multi-stage detection and segmentation of cerebral microbleeds
Aug 05, 2021



Abstract:Cerebral microbleeds are small, dark, round lesions that can be visualised on T2*-weighted MRI or other sequences sensitive to susceptibility effects. In this work, we propose a multi-stage approach to both microbleed detection and segmentation. First, possible microbleed locations are detected with a Mask R-CNN technique. Second, at each possible microbleed location, a simple U-Net performs the final segmentation. This work used the 72 subjects as training data provided by the "Where is VALDO?" challenge of MICCAI 2021.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge