Kenneth G. A. Gilhuijs
Multi-modal volumetric concept activation to explain detection and classification of metastatic prostate cancer on PSMA-PET/CT
Aug 04, 2022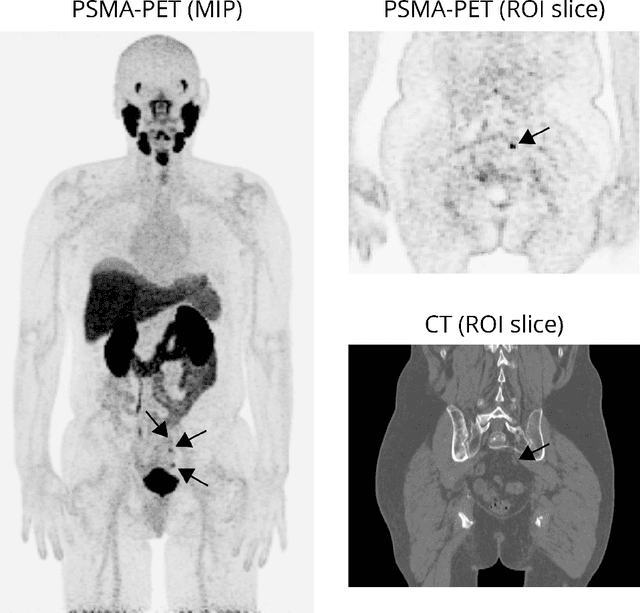

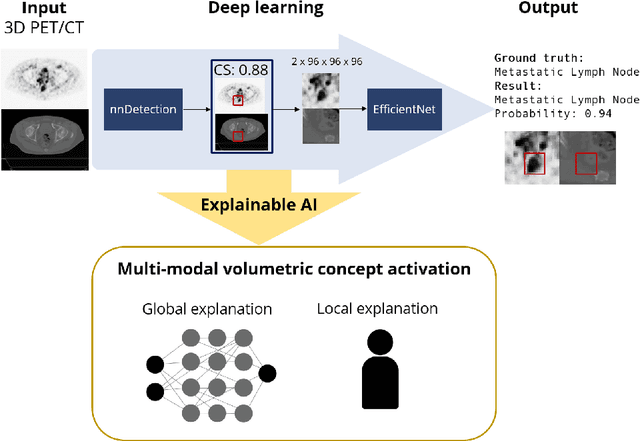
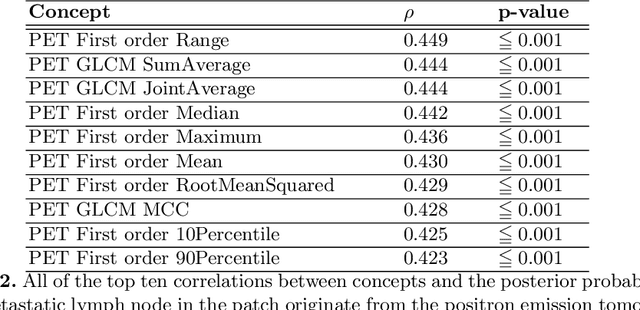
Abstract:Explainable artificial intelligence (XAI) is increasingly used to analyze the behavior of neural networks. Concept activation uses human-interpretable concepts to explain neural network behavior. This study aimed at assessing the feasibility of regression concept activation to explain detection and classification of multi-modal volumetric data. Proof-of-concept was demonstrated in metastatic prostate cancer patients imaged with positron emission tomography/computed tomography (PET/CT). Multi-modal volumetric concept activation was used to provide global and local explanations. Sensitivity was 80% at 1.78 false positive per patient. Global explanations showed that detection focused on CT for anatomical location and on PET for its confidence in the detection. Local explanations showed promise to aid in distinguishing true positives from false positives. Hence, this study demonstrated feasibility to explain detection and classification of multi-modal volumetric data using regression concept activation.
Explainable artificial intelligence (XAI) in deep learning-based medical image analysis
Jul 22, 2021



Abstract:With an increase in deep learning-based methods, the call for explainability of such methods grows, especially in high-stakes decision making areas such as medical image analysis. This survey presents an overview of eXplainable Artificial Intelligence (XAI) used in deep learning-based medical image analysis. A framework of XAI criteria is introduced to classify deep learning-based medical image analysis methods. Papers on XAI techniques in medical image analysis are then surveyed and categorized according to the framework and according to anatomical location. The paper concludes with an outlook of future opportunities for XAI in medical image analysis.
Interpretable deep learning regression for breast density estimation on MRI
Dec 08, 2020Abstract:Breast density, which is the ratio between fibroglandular tissue (FGT) and total breast volume, can be assessed qualitatively by radiologists and quantitatively by computer algorithms. These algorithms often rely on segmentation of breast and FGT volume. In this study, we propose a method to directly assess breast density on MRI, and provide interpretations of these assessments. We assessed breast density in 506 patients with breast cancer using a regression convolutional neural network (CNN). The input for the CNN were slices of breast MRI of 128 x 128 voxels, and the output was a continuous density value between 0 (fatty breast) and 1 (dense breast). We used 350 patients to train the CNN, 75 for validation, and 81 for independent testing. We investigated why the CNN came to its predicted density using Deep SHapley Additive exPlanations (SHAP). The density predicted by the CNN on the testing set was significantly correlated with the ground truth densities (N = 81 patients, Spearman's rho = 0.86, P < 0.001). When inspecting what the CNN based its predictions on, we found that voxels in FGT commonly had positive SHAP-values, voxels in fatty tissue commonly had negative SHAP-values, and voxels in non-breast tissue commonly had SHAP-values near zero. This means that the prediction of density is based on the structures we expect it to be based on, namely FGT and fatty tissue. To conclude, we presented an interpretable deep learning regression method for breast density estimation on MRI with promising results.
Response monitoring of breast cancer on DCE-MRI using convolutional neural network-generated seed points and constrained volume growing
Nov 22, 2018Abstract:Response of breast cancer to neoadjuvant chemotherapy (NAC) can be monitored using the change in visible tumor on magnetic resonance imaging (MRI). In our current workflow, seed points are manually placed in areas of enhancement likely to contain cancer. A constrained volume growing method uses these manually placed seed points as input and generates a tumor segmentation. This method is rigorously validated using complete pathological embedding. In this study, we propose to exploit deep learning for fast and automatic seed point detection, replacing manual seed point placement in our existing and well-validated workflow. The seed point generator was developed in early breast cancer patients with pathology-proven segmentations (N=100), operated shortly after MRI. It consisted of an ensemble of three independently trained fully convolutional dilated neural networks that classified breast voxels as tumor or non-tumor. Subsequently, local maxima were used as seed points for volume growing in patients receiving NAC (N=10). The percentage of tumor volume change was evaluated against semi-automatic segmentations. The primary cancer was localized in 95% of the tumors at the cost of 0.9 false positive per patient. False positives included focally enhancing regions of unknown origin and parts of the intramammary blood vessels. Volume growing from the seed points showed a median tumor volume decrease of 70% (interquartile range: 50%-77%), comparable to the semi-automatic segmentations (median: 70%, interquartile range 23%-76%). To conclude, a fast and automatic seed point generator was developed, fully automating a well-validated semi-automatic workflow for response monitoring of breast cancer to neoadjuvant chemotherapy.
Deep Learning for Multi-Task Medical Image Segmentation in Multiple Modalities
Apr 11, 2017


Abstract:Automatic segmentation of medical images is an important task for many clinical applications. In practice, a wide range of anatomical structures are visualised using different imaging modalities. In this paper, we investigate whether a single convolutional neural network (CNN) can be trained to perform different segmentation tasks. A single CNN is trained to segment six tissues in MR brain images, the pectoral muscle in MR breast images, and the coronary arteries in cardiac CTA. The CNN therefore learns to identify the imaging modality, the visualised anatomical structures, and the tissue classes. For each of the three tasks (brain MRI, breast MRI and cardiac CTA), this combined training procedure resulted in a segmentation performance equivalent to that of a CNN trained specifically for that task, demonstrating the high capacity of CNN architectures. Hence, a single system could be used in clinical practice to automatically perform diverse segmentation tasks without task-specific training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge