Han Yuan
Empowering Small Language Models with Factual Hallucination-Aware Reasoning for Financial Classification
Jan 04, 2026Abstract:Small language models (SLMs) are increasingly used for financial classification due to their fast inference and local deployability. However, compared with large language models, SLMs are more prone to factual hallucinations in reasoning and exhibit weaker classification performance. This raises a natural question: Can mitigating factual hallucinations improve SLMs' financial classification? To address this, we propose a three-step pipeline named AAAI (Association Identification, Automated Detection, and Adaptive Inference). Experiments on three representative SLMs reveal that: (1) factual hallucinations are positively correlated with misclassifications; (2) encoder-based verifiers effectively detect factual hallucinations; and (3) incorporating feedback on factual errors enables SLMs' adaptive inference that enhances classification performance. We hope this pipeline contributes to trustworthy and effective applications of SLMs in finance.
MKG-Rank: Enhancing Large Language Models with Knowledge Graph for Multilingual Medical Question Answering
Mar 21, 2025Abstract:Large Language Models (LLMs) have shown remarkable progress in medical question answering (QA), yet their effectiveness remains predominantly limited to English due to imbalanced multilingual training data and scarce medical resources for low-resource languages. To address this critical language gap in medical QA, we propose Multilingual Knowledge Graph-based Retrieval Ranking (MKG-Rank), a knowledge graph-enhanced framework that enables English-centric LLMs to perform multilingual medical QA. Through a word-level translation mechanism, our framework efficiently integrates comprehensive English-centric medical knowledge graphs into LLM reasoning at a low cost, mitigating cross-lingual semantic distortion and achieving precise medical QA across language barriers. To enhance efficiency, we introduce caching and multi-angle ranking strategies to optimize the retrieval process, significantly reducing response times and prioritizing relevant medical knowledge. Extensive evaluations on multilingual medical QA benchmarks across Chinese, Japanese, Korean, and Swahili demonstrate that MKG-Rank consistently outperforms zero-shot LLMs, achieving maximum 35.03% increase in accuracy, while maintaining an average retrieval time of only 0.0009 seconds.
Extract, Match, and Score: An Evaluation Paradigm for Long Question-context-answer Triplets in Financial Analysis
Mar 20, 2025Abstract:The rapid advancement of large language models (LLMs) has sparked widespread adoption across diverse applications, making robust evaluation frameworks crucial for assessing their performance. While conventional evaluation metrics remain applicable for shorter texts, their efficacy diminishes when evaluating the quality of long-form answers. This limitation is particularly critical in real-world scenarios involving extended questions, extensive context, and long-form answers, such as financial analysis or regulatory compliance. In this paper, we use a practical financial use case to illustrate applications that handle "long question-context-answer triplets". We construct a real-world financial dataset comprising long triplets and demonstrate the inadequacies of traditional metrics. To address this, we propose an effective Extract, Match, and Score (EMS) evaluation approach tailored to the complexities of long-form LLMs' outputs, providing practitioners with a reliable methodology for assessing LLMs' performance in complex real-world scenarios.
Exploring the Reliability of Self-explanation and its Relationship with Classification in Language Model-driven Financial Analysis
Mar 20, 2025Abstract:Language models (LMs) have exhibited exceptional versatility in reasoning and in-depth financial analysis through their proprietary information processing capabilities. Previous research focused on evaluating classification performance while often overlooking explainability or pre-conceived that refined explanation corresponds to higher classification accuracy. Using a public dataset in finance domain, we quantitatively evaluated self-explanations by LMs, focusing on their factuality and causality. We identified the statistically significant relationship between the accuracy of classifications and the factuality or causality of self-explanations. Our study built an empirical foundation for approximating classification confidence through self-explanations and for optimizing classification via proprietary reasoning.
GraphCheck: Breaking Long-Term Text Barriers with Extracted Knowledge Graph-Powered Fact-Checking
Feb 23, 2025Abstract:Large language models (LLMs) are widely used, but they often generate subtle factual errors, especially in long-form text. These errors are fatal in some specialized domains such as medicine. Existing fact-checking with grounding documents methods face two main challenges: (1) they struggle to understand complex multihop relations in long documents, often overlooking subtle factual errors; (2) most specialized methods rely on pairwise comparisons, requiring multiple model calls, leading to high resource and computational costs. To address these challenges, we propose \textbf{\textit{GraphCheck}}, a fact-checking framework that uses extracted knowledge graphs to enhance text representation. Graph Neural Networks further process these graphs as a soft prompt, enabling LLMs to incorporate structured knowledge more effectively. Enhanced with graph-based reasoning, GraphCheck captures multihop reasoning chains which are often overlooked by existing methods, enabling precise and efficient fact-checking in a single inference call. Experimental results on seven benchmarks spanning both general and medical domains demonstrate a 6.1\% overall improvement over baseline models. Notably, GraphCheck outperforms existing specialized fact-checkers and achieves comparable performance with state-of-the-art LLMs, such as DeepSeek-V3 and OpenAI-o1, with significantly fewer parameters.
Clinical Domain Knowledge-Derived Template Improves Post Hoc AI Explanations in Pneumothorax Classification
Mar 26, 2024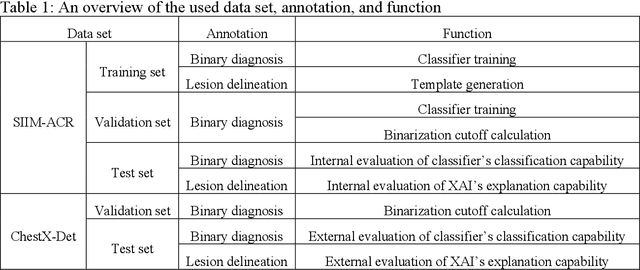

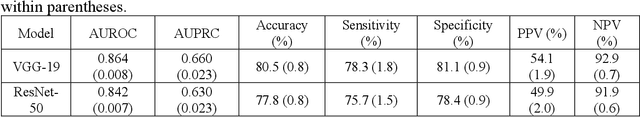

Abstract:Background: Pneumothorax is an acute thoracic disease caused by abnormal air collection between the lungs and chest wall. To address the opaqueness often associated with deep learning (DL) models, explainable artificial intelligence (XAI) methods have been introduced to outline regions related to pneumothorax diagnoses made by DL models. However, these explanations sometimes diverge from actual lesion areas, highlighting the need for further improvement. Method: We propose a template-guided approach to incorporate the clinical knowledge of pneumothorax into model explanations generated by XAI methods, thereby enhancing the quality of these explanations. Utilizing one lesion delineation created by radiologists, our approach first generates a template that represents potential areas of pneumothorax occurrence. This template is then superimposed on model explanations to filter out extraneous explanations that fall outside the template's boundaries. To validate its efficacy, we carried out a comparative analysis of three XAI methods with and without our template guidance when explaining two DL models in two real-world datasets. Results: The proposed approach consistently improved baseline XAI methods across twelve benchmark scenarios built on three XAI methods, two DL models, and two datasets. The average incremental percentages, calculated by the performance improvements over the baseline performance, were 97.8% in Intersection over Union (IoU) and 94.1% in Dice Similarity Coefficient (DSC) when comparing model explanations and ground-truth lesion areas. Conclusions: In the context of pneumothorax diagnoses, we proposed a template-guided approach for improving AI explanations. We anticipate that our template guidance will forge a fresh approach to elucidating AI models by integrating clinical domain expertise.
Efficient scene text image super-resolution with semantic guidance
Mar 20, 2024



Abstract:Scene text image super-resolution has significantly improved the accuracy of scene text recognition. However, many existing methods emphasize performance over efficiency and ignore the practical need for lightweight solutions in deployment scenarios. Faced with the issues, our work proposes an efficient framework called SGENet to facilitate deployment on resource-limited platforms. SGENet contains two branches: super-resolution branch and semantic guidance branch. We apply a lightweight pre-trained recognizer as a semantic extractor to enhance the understanding of text information. Meanwhile, we design the visual-semantic alignment module to achieve bidirectional alignment between image features and semantics, resulting in the generation of highquality prior guidance. We conduct extensive experiments on benchmark dataset, and the proposed SGENet achieves excellent performance with fewer computational costs. Code is available at https://github.com/SijieLiu518/SGENet
Foundation Model Makes Clustering a Better Initialization for Active Learning
Feb 04, 2024



Abstract:Active learning selects the most informative samples from the unlabeled dataset to annotate in the context of a limited annotation budget. While numerous methods have been proposed for subsequent sample selection based on an initialized model, scant attention has been paid to the indispensable phase of active learning: selecting samples for model initialization. Most of the previous studies resort to random sampling or naive clustering. However, random sampling is prone to fluctuation, and naive clustering suffers from convergence speed, particularly when dealing with high-dimensional data such as imaging data. In this work, we propose to integrate foundation models with clustering methods to select samples for active learning initialization. Foundation models refer to those trained on massive datasets by the self-supervised paradigm and capable of generating informative and compacted embeddings for various downstream tasks. Leveraging these embeddings to replace raw features such as pixel values, clustering quickly converges and identifies better initial samples. For a comprehensive comparison, we included a classic ImageNet-supervised model to acquire embeddings. Experiments on two clinical tasks of image classification and segmentation demonstrated that foundation model-based clustering efficiently pinpointed informative initial samples, leading to models showcasing enhanced performance than the baseline methods. We envisage that this study provides an effective paradigm for future active learning.
Leveraging Anatomical Constraints with Uncertainty for Pneumothorax Segmentation
Nov 26, 2023



Abstract:Pneumothorax is a medical emergency caused by abnormal accumulation of air in the pleural space - the potential space between the lungs and chest wall. On 2D chest radiographs, pneumothorax occurs within the thoracic cavity and outside of the mediastinum and we refer to this area as "lung+ space". While deep learning (DL) has increasingly been utilized to segment pneumothorax lesions in chest radiographs, many existing DL models employ an end-to-end approach. These models directly map chest radiographs to clinician-annotated lesion areas, often neglecting the vital domain knowledge that pneumothorax is inherently location-sensitive. We propose a novel approach that incorporates the lung+ space as a constraint during DL model training for pneumothorax segmentation on 2D chest radiographs. To circumvent the need for additional annotations and to prevent potential label leakage on the target task, our method utilizes external datasets and an auxiliary task of lung segmentation. This approach generates a specific constraint of lung+ space for each chest radiograph. Furthermore, we have incorporated a discriminator to eliminate unreliable constraints caused by the domain shift between the auxiliary and target datasets. Our results demonstrated significant improvements, with average performance gains of 4.6%, 3.6%, and 3.3% regarding Intersection over Union (IoU), Dice Similarity Coefficient (DSC), and Hausdorff Distance (HD). Our research underscores the significance of incorporating medical domain knowledge about the location-specific nature of pneumothorax to enhance DL-based lesion segmentation.
FedScore: A privacy-preserving framework for federated scoring system development
Mar 01, 2023

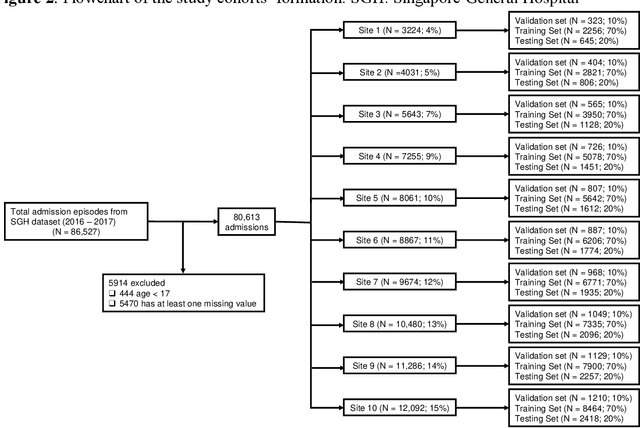

Abstract:We propose FedScore, a privacy-preserving federated learning framework for scoring system generation across multiple sites to facilitate cross-institutional collaborations. The FedScore framework includes five modules: federated variable ranking, federated variable transformation, federated score derivation, federated model selection and federated model evaluation. To illustrate usage and assess FedScore's performance, we built a hypothetical global scoring system for mortality prediction within 30 days after a visit to an emergency department using 10 simulated sites divided from a tertiary hospital in Singapore. We employed a pre-existing score generator to construct 10 local scoring systems independently at each site and we also developed a scoring system using centralized data for comparison. We compared the acquired FedScore model's performance with that of other scoring models using the receiver operating characteristic (ROC) analysis. The FedScore model achieved an average area under the curve (AUC) value of 0.763 across all sites, with a standard deviation (SD) of 0.020. We also calculated the average AUC values and SDs for each local model, and the FedScore model showed promising accuracy and stability with a high average AUC value which was closest to the one of the pooled model and SD which was lower than that of most local models. This study demonstrates that FedScore is a privacy-preserving scoring system generator with potentially good generalizability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge