Seyed Ehsan Saffari
Handling missing values in healthcare data: A systematic review of deep learning-based imputation techniques
Oct 15, 2022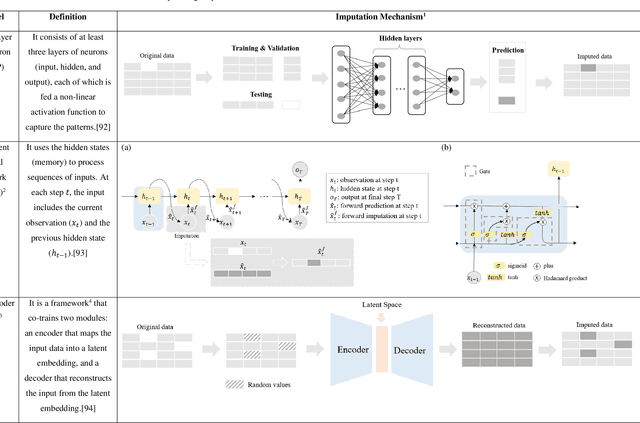
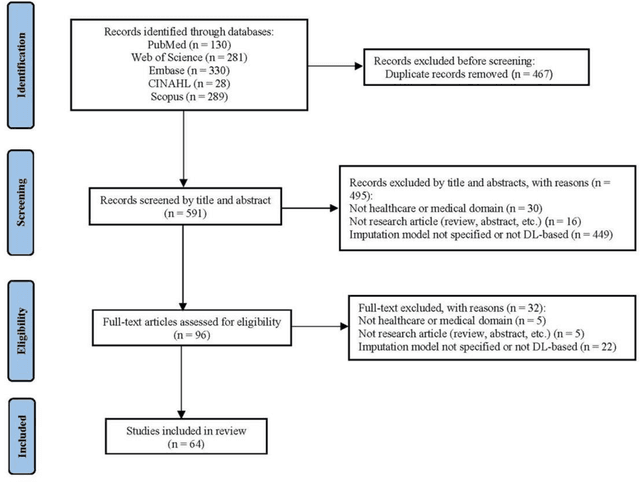
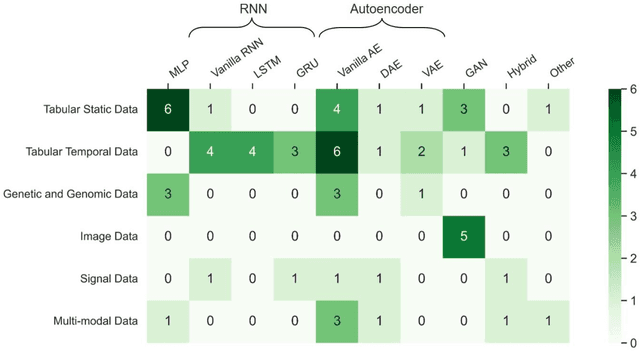
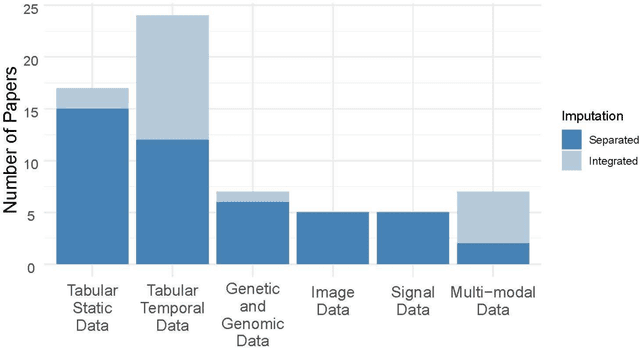
Abstract:Objective: The proper handling of missing values is critical to delivering reliable estimates and decisions, especially in high-stakes fields such as clinical research. The increasing diversity and complexity of data have led many researchers to develop deep learning (DL)-based imputation techniques. We conducted a systematic review to evaluate the use of these techniques, with a particular focus on data types, aiming to assist healthcare researchers from various disciplines in dealing with missing values. Methods: We searched five databases (MEDLINE, Web of Science, Embase, CINAHL, and Scopus) for articles published prior to August 2021 that applied DL-based models to imputation. We assessed selected publications from four perspectives: health data types, model backbone (i.e., main architecture), imputation strategies, and comparison with non-DL-based methods. Based on data types, we created an evidence map to illustrate the adoption of DL models. Results: We included 64 articles, of which tabular static (26.6%, 17/64) and temporal data (37.5%, 24/64) were the most frequently investigated. We found that model backbone(s) differed among data types as well as the imputation strategy. The "integrated" strategy, that is, the imputation task being solved concurrently with downstream tasks, was popular for tabular temporal (50%, 12/24) and multi-modal data (71.4%, 5/7), but limited for other data types. Moreover, DL-based imputation methods yielded better imputation accuracy in most studies, compared with non-DL-based methods. Conclusion: DL-based imputation models can be customized based on data type, addressing the corresponding missing patterns, and its associated "integrated" strategy can enhance the efficacy of imputation, especially in scenarios where data is complex. Future research may focus on the portability and fairness of DL-based models for healthcare data imputation.
AutoScore-Ordinal: An interpretable machine learning framework for generating scoring models for ordinal outcomes
Feb 17, 2022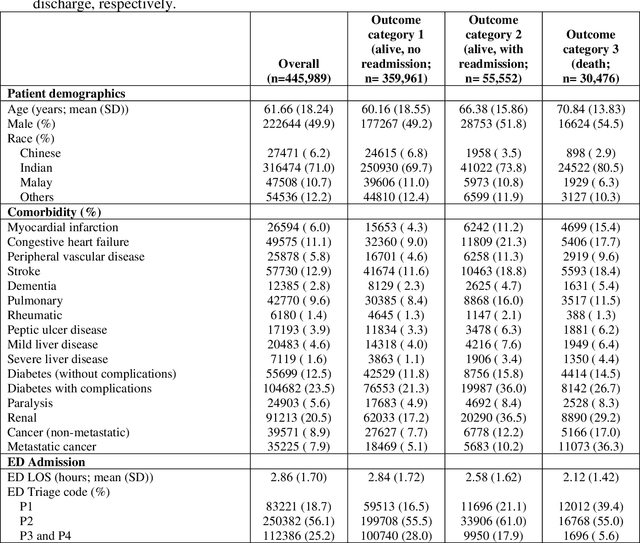
Abstract:Background: Risk prediction models are useful tools in clinical decision-making which help with risk stratification and resource allocations and may lead to a better health care for patients. AutoScore is a machine learning-based automatic clinical score generator for binary outcomes. This study aims to expand the AutoScore framework to provide a tool for interpretable risk prediction for ordinal outcomes. Methods: The AutoScore-Ordinal framework is generated using the same 6 modules of the original AutoScore algorithm including variable ranking, variable transformation, score derivation (from proportional odds models), model selection, score fine-tuning, and model evaluation. To illustrate the AutoScore-Ordinal performance, the method was conducted on electronic health records data from the emergency department at Singapore General Hospital over 2008 to 2017. The model was trained on 70% of the data, validated on 10% and tested on the remaining 20%. Results: This study included 445,989 inpatient cases, where the distribution of the ordinal outcome was 80.7% alive without 30-day readmission, 12.5% alive with 30-day readmission, and 6.8% died inpatient or by day 30 post discharge. Two point-based risk prediction models were developed using two sets of 8 predictor variables identified by the flexible variable selection procedure. The two models indicated reasonably good performance measured by mean area under the receiver operating characteristic curve (0.785 and 0.793) and generalized c-index (0.737 and 0.760), which were comparable to alternative models. Conclusion: AutoScore-Ordinal provides an automated and easy-to-use framework for development and validation of risk prediction models for ordinal outcomes, which can systematically identify potential predictors from high-dimensional data.
AutoScore-Imbalance: An interpretable machine learning tool for development of clinical scores with rare events data
Jul 13, 2021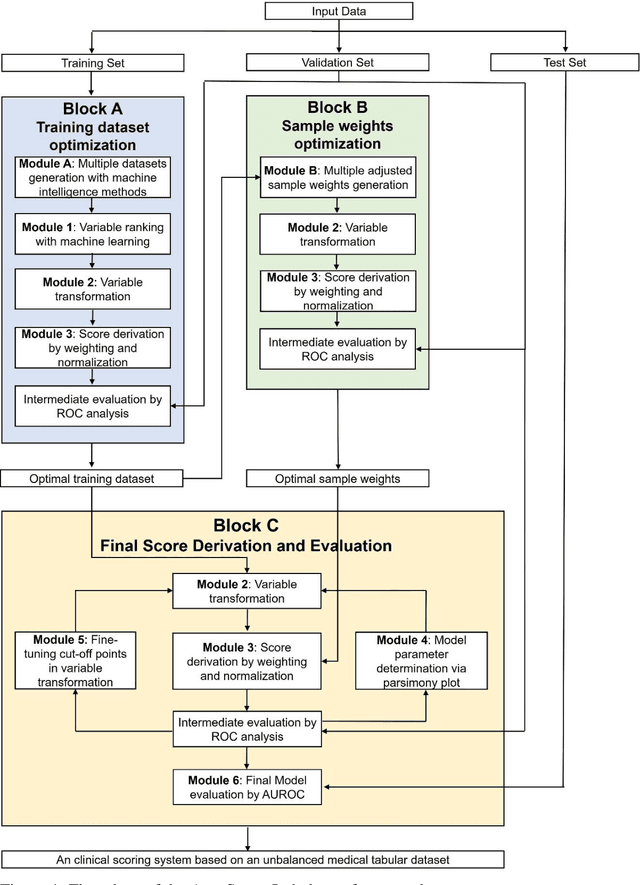
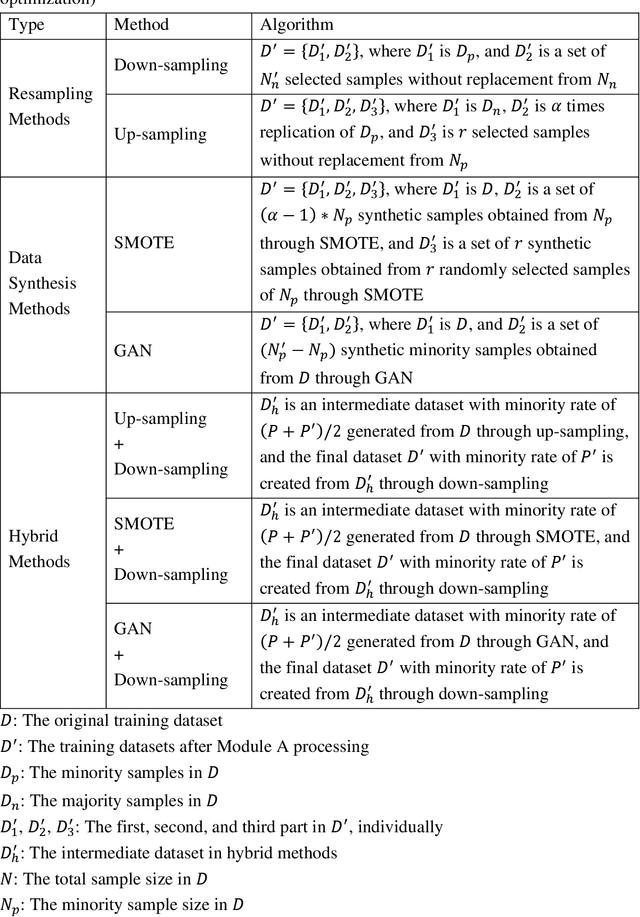
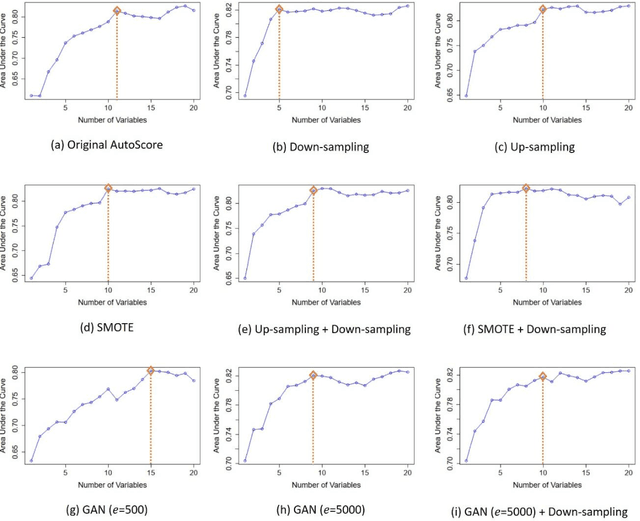
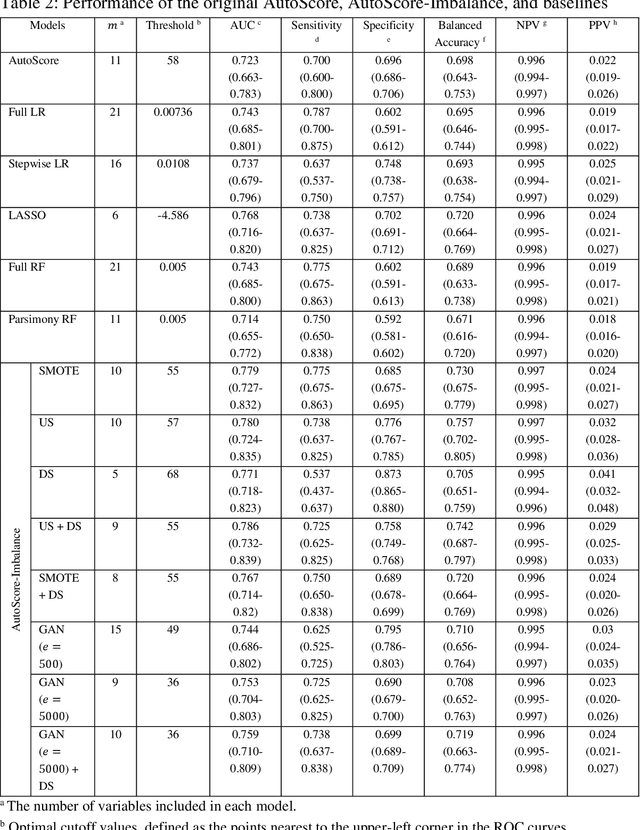
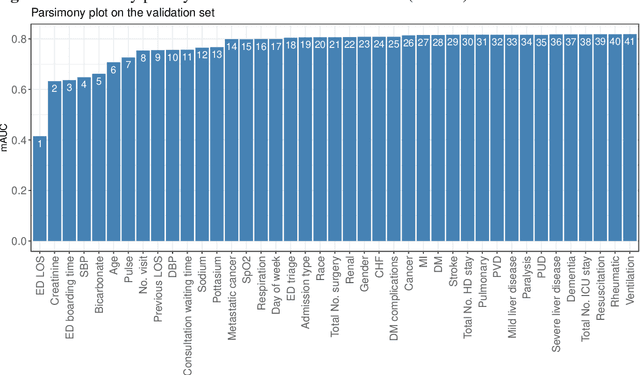
Abstract:Background: Medical decision-making impacts both individual and public health. Clinical scores are commonly used among a wide variety of decision-making models for determining the degree of disease deterioration at the bedside. AutoScore was proposed as a useful clinical score generator based on machine learning and a generalized linear model. Its current framework, however, still leaves room for improvement when addressing unbalanced data of rare events. Methods: Using machine intelligence approaches, we developed AutoScore-Imbalance, which comprises three components: training dataset optimization, sample weight optimization, and adjusted AutoScore. All scoring models were evaluated on the basis of their area under the curve (AUC) in the receiver operating characteristic analysis and balanced accuracy (i.e., mean value of sensitivity and specificity). By utilizing a publicly accessible dataset from Beth Israel Deaconess Medical Center, we assessed the proposed model and baseline approaches in the prediction of inpatient mortality. Results: AutoScore-Imbalance outperformed baselines in terms of AUC and balanced accuracy. The nine-variable AutoScore-Imbalance sub-model achieved the highest AUC of 0.786 (0.732-0.839) while the eleven-variable original AutoScore obtained an AUC of 0.723 (0.663-0.783), and the logistic regression with 21 variables obtained an AUC of 0.743 (0.685-0.800). The AutoScore-Imbalance sub-model (using down-sampling algorithm) yielded an AUC of 0. 0.771 (0.718-0.823) with only five variables, demonstrating a good balance between performance and variable sparsity. Conclusions: The AutoScore-Imbalance tool has the potential to be applied to highly unbalanced datasets to gain further insight into rare medical events and to facilitate real-world clinical decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge