Benjamin Alan Goldstein
A roadmap to fair and trustworthy prediction model validation in healthcare
Apr 07, 2023Abstract:A prediction model is most useful if it generalizes beyond the development data with external validations, but to what extent should it generalize remains unclear. In practice, prediction models are externally validated using data from very different settings, including populations from other health systems or countries, with predictably poor results. This may not be a fair reflection of the performance of the model which was designed for a specific target population or setting, and may be stretching the expected model generalizability. To address this, we suggest to externally validate a model using new data from the target population to ensure clear implications of validation performance on model reliability, whereas model generalizability to broader settings should be carefully investigated during model development instead of explored post-hoc. Based on this perspective, we propose a roadmap that facilitates the development and application of reliable, fair, and trustworthy artificial intelligence prediction models.
Shapley variable importance clouds for interpretable machine learning
Oct 06, 2021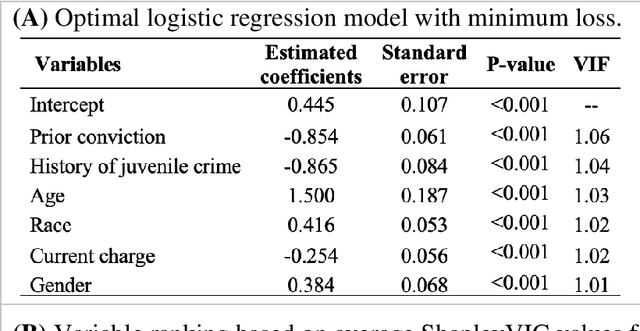
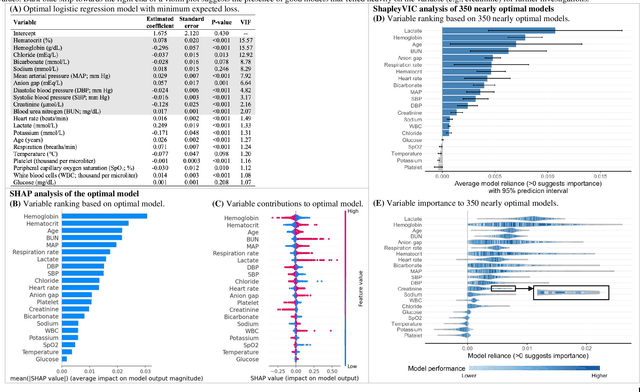
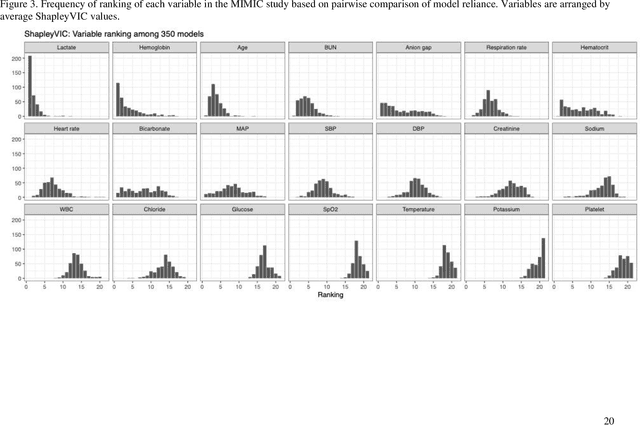
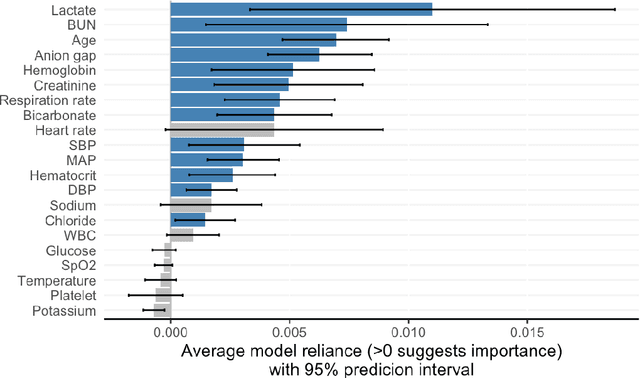
Abstract:Interpretable machine learning has been focusing on explaining final models that optimize performance. The current state-of-the-art is the Shapley additive explanations (SHAP) that locally explains variable impact on individual predictions, and it is recently extended for a global assessment across the dataset. Recently, Dong and Rudin proposed to extend the investigation to models from the same class as the final model that are "good enough", and identified a previous overclaim of variable importance based on a single model. However, this method does not directly integrate with existing Shapley-based interpretations. We close this gap by proposing a Shapley variable importance cloud that pools information across good models to avoid biased assessments in SHAP analyses of final models, and communicate the findings via novel visualizations. We demonstrate the additional insights gain compared to conventional explanations and Dong and Rudin's method using criminal justice and electronic medical records data.
AutoScore-Imbalance: An interpretable machine learning tool for development of clinical scores with rare events data
Jul 13, 2021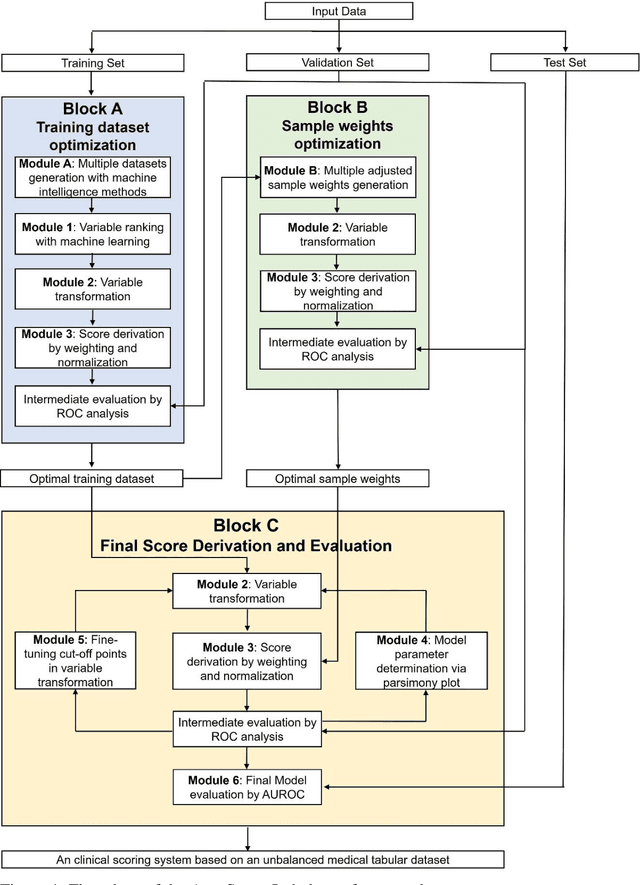
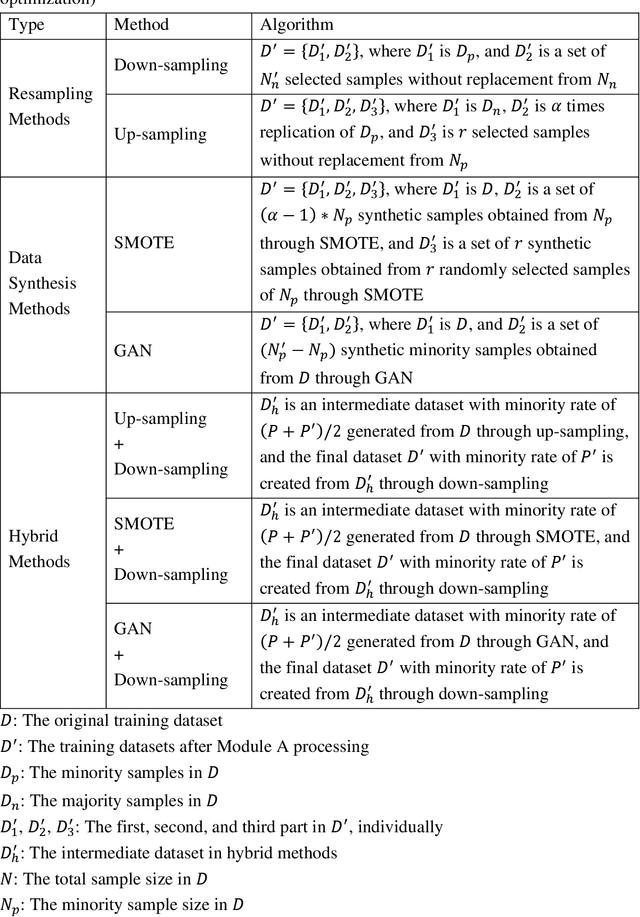
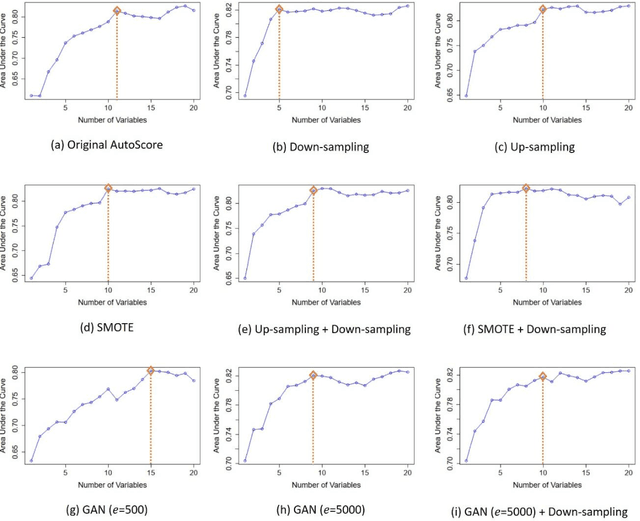
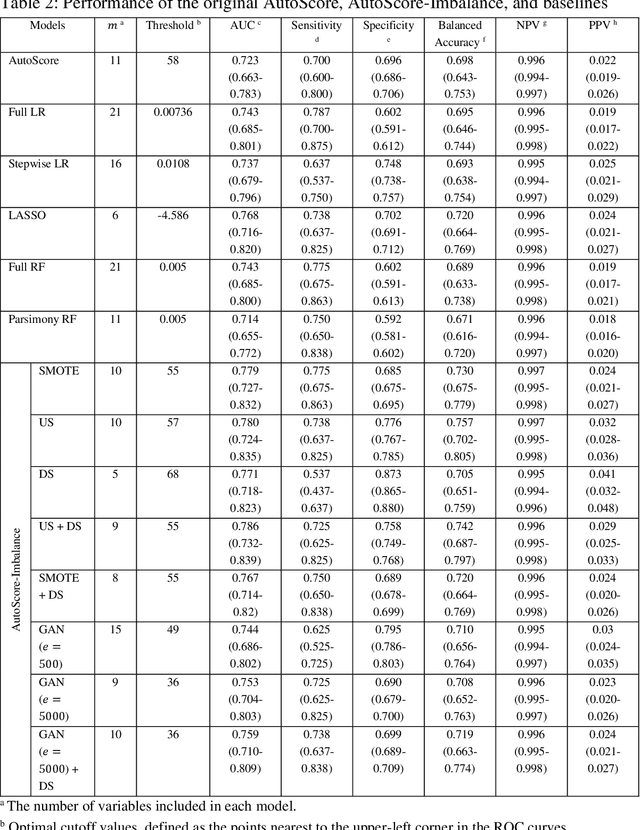
Abstract:Background: Medical decision-making impacts both individual and public health. Clinical scores are commonly used among a wide variety of decision-making models for determining the degree of disease deterioration at the bedside. AutoScore was proposed as a useful clinical score generator based on machine learning and a generalized linear model. Its current framework, however, still leaves room for improvement when addressing unbalanced data of rare events. Methods: Using machine intelligence approaches, we developed AutoScore-Imbalance, which comprises three components: training dataset optimization, sample weight optimization, and adjusted AutoScore. All scoring models were evaluated on the basis of their area under the curve (AUC) in the receiver operating characteristic analysis and balanced accuracy (i.e., mean value of sensitivity and specificity). By utilizing a publicly accessible dataset from Beth Israel Deaconess Medical Center, we assessed the proposed model and baseline approaches in the prediction of inpatient mortality. Results: AutoScore-Imbalance outperformed baselines in terms of AUC and balanced accuracy. The nine-variable AutoScore-Imbalance sub-model achieved the highest AUC of 0.786 (0.732-0.839) while the eleven-variable original AutoScore obtained an AUC of 0.723 (0.663-0.783), and the logistic regression with 21 variables obtained an AUC of 0.743 (0.685-0.800). The AutoScore-Imbalance sub-model (using down-sampling algorithm) yielded an AUC of 0. 0.771 (0.718-0.823) with only five variables, demonstrating a good balance between performance and variable sparsity. Conclusions: The AutoScore-Imbalance tool has the potential to be applied to highly unbalanced datasets to gain further insight into rare medical events and to facilitate real-world clinical decision-making.
AutoScore-Survival: Developing interpretable machine learning-based time-to-event scores with right-censored survival data
Jun 13, 2021



Abstract:Scoring systems are highly interpretable and widely used to evaluate time-to-event outcomes in healthcare research. However, existing time-to-event scores are predominantly created ad-hoc using a few manually selected variables based on clinician's knowledge, suggesting an unmet need for a robust and efficient generic score-generating method. AutoScore was previously developed as an interpretable machine learning score generator, integrated both machine learning and point-based scores in the strong discriminability and accessibility. We have further extended it to time-to-event data and developed AutoScore-Survival, for automatically generating time-to-event scores with right-censored survival data. Random survival forest provides an efficient solution for selecting variables, and Cox regression was used for score weighting. We illustrated our method in a real-life study of 90-day mortality of patients in intensive care units and compared its performance with survival models (i.e., Cox) and the random survival forest. The AutoScore-Survival-derived scoring model was more parsimonious than survival models built using traditional variable selection methods (e.g., penalized likelihood approach and stepwise variable selection), and its performance was comparable to survival models using the same set of variables. Although AutoScore-Survival achieved a comparable integrated area under the curve of 0.782 (95% CI: 0.767-0.794), the integer-valued time-to-event scores generated are favorable in clinical applications because they are easier to compute and interpret. Our proposed AutoScore-Survival provides an automated, robust and easy-to-use machine learning-based clinical score generator to studies of time-to-event outcomes. It provides a systematic guideline to facilitate the future development of time-to-event scores for clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge