Hossein Rouhizadeh
Toward Global Large Language Models in Medicine
Jan 05, 2026Abstract:Despite continuous advances in medical technology, the global distribution of health care resources remains uneven. The development of large language models (LLMs) has transformed the landscape of medicine and holds promise for improving health care quality and expanding access to medical information globally. However, existing LLMs are primarily trained on high-resource languages, limiting their applicability in global medical scenarios. To address this gap, we constructed GlobMed, a large multilingual medical dataset, containing over 500,000 entries spanning 12 languages, including four low-resource languages. Building on this, we established GlobMed-Bench, which systematically assesses 56 state-of-the-art proprietary and open-weight LLMs across multiple multilingual medical tasks, revealing significant performance disparities across languages, particularly for low-resource languages. Additionally, we introduced GlobMed-LLMs, a suite of multilingual medical LLMs trained on GlobMed, with parameters ranging from 1.7B to 8B. GlobMed-LLMs achieved an average performance improvement of over 40% relative to baseline models, with a more than threefold increase in performance on low-resource languages. Together, these resources provide an important foundation for advancing the equitable development and application of LLMs globally, enabling broader language communities to benefit from technological advances.
MKG-Rank: Enhancing Large Language Models with Knowledge Graph for Multilingual Medical Question Answering
Mar 21, 2025Abstract:Large Language Models (LLMs) have shown remarkable progress in medical question answering (QA), yet their effectiveness remains predominantly limited to English due to imbalanced multilingual training data and scarce medical resources for low-resource languages. To address this critical language gap in medical QA, we propose Multilingual Knowledge Graph-based Retrieval Ranking (MKG-Rank), a knowledge graph-enhanced framework that enables English-centric LLMs to perform multilingual medical QA. Through a word-level translation mechanism, our framework efficiently integrates comprehensive English-centric medical knowledge graphs into LLM reasoning at a low cost, mitigating cross-lingual semantic distortion and achieving precise medical QA across language barriers. To enhance efficiency, we introduce caching and multi-angle ranking strategies to optimize the retrieval process, significantly reducing response times and prioritizing relevant medical knowledge. Extensive evaluations on multilingual medical QA benchmarks across Chinese, Japanese, Korean, and Swahili demonstrate that MKG-Rank consistently outperforms zero-shot LLMs, achieving maximum 35.03% increase in accuracy, while maintaining an average retrieval time of only 0.0009 seconds.
DS4DH at #SMM4H 2023: Zero-Shot Adverse Drug Events Normalization using Sentence Transformers and Reciprocal-Rank Fusion
Aug 15, 2023

Abstract:This paper outlines the performance evaluation of a system for adverse drug event normalization, developed by the Data Science for Digital Health group for the Social Media Mining for Health Applications 2023 shared task 5. Shared task 5 targeted the normalization of adverse drug event mentions in Twitter to standard concepts from the Medical Dictionary for Regulatory Activities terminology. Our system hinges on a two-stage approach: BERT fine-tuning for entity recognition, followed by zero-shot normalization using sentence transformers and reciprocal-rank fusion. The approach yielded a precision of 44.9%, recall of 40.5%, and an F1-score of 42.6%. It outperformed the median performance in shared task 5 by 10% and demonstrated the highest performance among all participants. These results substantiate the effectiveness of our approach and its potential application for adverse drug event normalization in the realm of social media text mining.
Efficient Joint Learning for Clinical Named Entity Recognition and Relation Extraction Using Fourier Networks: A Use Case in Adverse Drug Events
Feb 08, 2023



Abstract:Current approaches for clinical information extraction are inefficient in terms of computational costs and memory consumption, hindering their application to process large-scale electronic health records (EHRs). We propose an efficient end-to-end model, the Joint-NER-RE-Fourier (JNRF), to jointly learn the tasks of named entity recognition and relation extraction for documents of variable length. The architecture uses positional encoding and unitary batch sizes to process variable length documents and uses a weight-shared Fourier network layer for low-complexity token mixing. Finally, we reach the theoretical computational complexity lower bound for relation extraction using a selective pooling strategy and distance-aware attention weights with trainable polynomial distance functions. We evaluated the JNRF architecture using the 2018 N2C2 ADE benchmark to jointly extract medication-related entities and relations in variable-length EHR summaries. JNRF outperforms rolling window BERT with selective pooling by 0.42%, while being twice as fast to train. Compared to state-of-the-art BiLSTM-CRF architectures on the N2C2 ADE benchmark, results show that the proposed approach trains 22 times faster and reduces GPU memory consumption by 1.75 folds, with a reasonable performance tradeoff of 90%, without the use of external tools, hand-crafted rules or post-processing. Given the significant carbon footprint of deep learning models and the current energy crises, these methods could support efficient and cleaner information extraction in EHRs and other types of large-scale document databases.
Persian-WSD-Corpus: A Sense Annotated Corpus for Persian All-words Word Sense Disambiguation
Jul 04, 2021

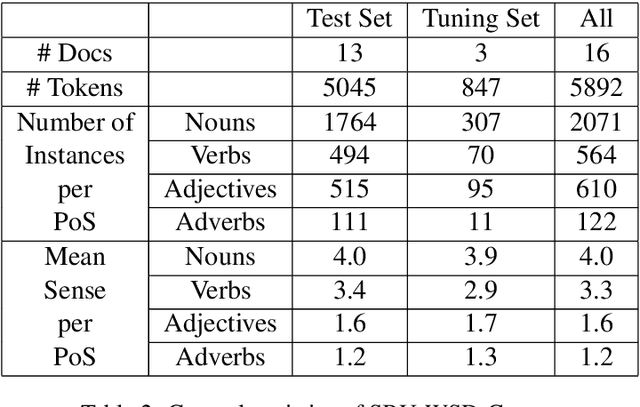
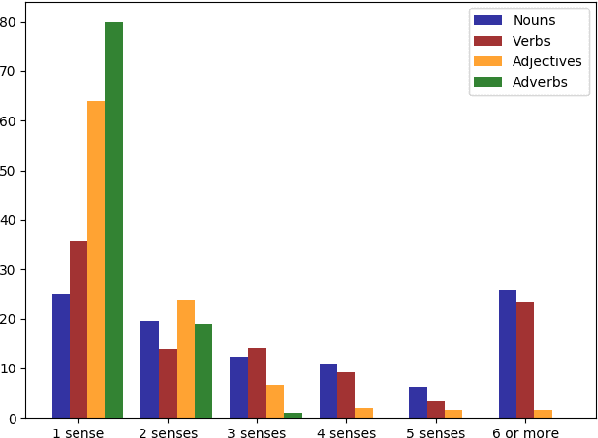
Abstract:Word Sense Disambiguation (WSD) is a long-standing task in Natural Language Processing(NLP) that aims to automatically identify the most relevant meaning of the words in a given context. Developing standard WSD test collections can be mentioned as an important prerequisite for developing and evaluating different WSD systems in the language of interest. Although many WSD test collections have been developed for a variety of languages, no standard All-words WSD benchmark is available for Persian. In this paper, we address this shortage for the Persian language by introducing SBU-WSD-Corpus, as the first standard test set for the Persian All-words WSD task. SBU-WSD-Corpus is manually annotated with senses from the Persian WordNet (FarsNet) sense inventory. To this end, three annotators used SAMP (a tool for sense annotation based on FarsNet lexical graph) to perform the annotation task. SBU-WSD-Corpus consists of 19 Persian documents in different domains such as Sports, Science, Arts, etc. It includes 5892 content words of Persian running text and 3371 manually sense annotated words (2073 nouns, 566 verbs, 610 adjectives, and 122 adverbs). Providing baselines for future studies on the Persian All-words WSD task, we evaluate several WSD models on SBU-WSD-Corpus. The corpus is publicly available at https://github.com/hrouhizadeh/SBU-WSD-Corpus.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge