Anthony Yazdani
ICU-TSB: A Benchmark for Temporal Patient Representation Learning for Unsupervised Stratification into Patient Cohorts
Jun 06, 2025Abstract:Patient stratification identifying clinically meaningful subgroups is essential for advancing personalized medicine through improved diagnostics and treatment strategies. Electronic health records (EHRs), particularly those from intensive care units (ICUs), contain rich temporal clinical data that can be leveraged for this purpose. In this work, we introduce ICU-TSB (Temporal Stratification Benchmark), the first comprehensive benchmark for evaluating patient stratification based on temporal patient representation learning using three publicly available ICU EHR datasets. A key contribution of our benchmark is a novel hierarchical evaluation framework utilizing disease taxonomies to measure the alignment of discovered clusters with clinically validated disease groupings. In our experiments with ICU-TSB, we compared statistical methods and several recurrent neural networks, including LSTM and GRU, for their ability to generate effective patient representations for subsequent clustering of patient trajectories. Our results demonstrate that temporal representation learning can rediscover clinically meaningful patient cohorts; nevertheless, it remains a challenging task, with v-measuring varying from up to 0.46 at the top level of the taxonomy to up to 0.40 at the lowest level. To further enhance the practical utility of our findings, we also evaluate multiple strategies for assigning interpretable labels to the identified clusters. The experiments and benchmark are fully reproducible and available at https://github.com/ds4dh/CBMS2025stratification.
GLiNER-biomed: A Suite of Efficient Models for Open Biomedical Named Entity Recognition
Apr 01, 2025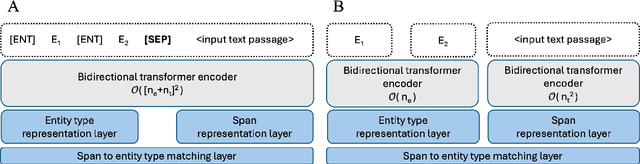



Abstract:Biomedical named entity recognition (NER) presents unique challenges due to specialized vocabularies, the sheer volume of entities, and the continuous emergence of novel entities. Traditional NER models, constrained by fixed taxonomies and human annotations, struggle to generalize beyond predefined entity types or efficiently adapt to emerging concepts. To address these issues, we introduce GLiNER-biomed, a domain-adapted suite of Generalist and Lightweight Model for NER (GLiNER) models specifically tailored for biomedical NER. In contrast to conventional approaches, GLiNER uses natural language descriptions to infer arbitrary entity types, enabling zero-shot recognition. Our approach first distills the annotation capabilities of large language models (LLMs) into a smaller, more efficient model, enabling the generation of high-coverage synthetic biomedical NER data. We subsequently train two GLiNER architectures, uni- and bi-encoder, at multiple scales to balance computational efficiency and recognition performance. Evaluations on several biomedical datasets demonstrate that GLiNER-biomed outperforms state-of-the-art GLiNER models in both zero- and few-shot scenarios, achieving 5.96% improvement in F1-score over the strongest baseline. Ablation studies highlight the effectiveness of our synthetic data generation strategy and emphasize the complementary benefits of synthetic biomedical pre-training combined with fine-tuning on high-quality general-domain annotations. All datasets, models, and training pipelines are publicly available at https://github.com/ds4dh/GLiNER-biomed.
CT-ADE: An Evaluation Benchmark for Adverse Drug Event Prediction from Clinical Trial Results
Apr 19, 2024Abstract:Adverse drug events (ADEs) significantly impact clinical research and public health, contributing to failures in clinical trials and leading to increased healthcare costs. The accurate prediction and management of ADEs are crucial for improving the development of safer, more effective medications, and enhancing patient outcomes. To support this effort, we introduce CT-ADE, a novel dataset compiled to enhance the predictive modeling of ADEs. Encompassing over 12,000 instances extracted from clinical trial results, the CT-ADE dataset integrates drug, patient population, and contextual information for multilabel ADE classification tasks in monopharmacy treatments, providing a comprehensive resource for developing advanced predictive models. To mirror the complex nature of ADEs, annotations are standardized at the system organ class level of the Medical Dictionary for Regulatory Activities (MedDRA) ontology. Preliminary analyses using baseline models have demonstrated promising results, achieving 73.33% F1 score and 81.54% balanced accuracy, highlighting CT-ADE's potential to advance ADE prediction. CT-ADE provides an essential tool for researchers aiming to leverage the power of artificial intelligence and machine learning to enhance patient safety and minimize the impact of ADEs on pharmaceutical research and development. Researchers interested in using the CT-ADE dataset can find all necessary resources at https://github.com/xxxx/xxxx.
DS4DH at #SMM4H 2023: Zero-Shot Adverse Drug Events Normalization using Sentence Transformers and Reciprocal-Rank Fusion
Aug 15, 2023

Abstract:This paper outlines the performance evaluation of a system for adverse drug event normalization, developed by the Data Science for Digital Health group for the Social Media Mining for Health Applications 2023 shared task 5. Shared task 5 targeted the normalization of adverse drug event mentions in Twitter to standard concepts from the Medical Dictionary for Regulatory Activities terminology. Our system hinges on a two-stage approach: BERT fine-tuning for entity recognition, followed by zero-shot normalization using sentence transformers and reciprocal-rank fusion. The approach yielded a precision of 44.9%, recall of 40.5%, and an F1-score of 42.6%. It outperformed the median performance in shared task 5 by 10% and demonstrated the highest performance among all participants. These results substantiate the effectiveness of our approach and its potential application for adverse drug event normalization in the realm of social media text mining.
Efficient Joint Learning for Clinical Named Entity Recognition and Relation Extraction Using Fourier Networks: A Use Case in Adverse Drug Events
Feb 08, 2023



Abstract:Current approaches for clinical information extraction are inefficient in terms of computational costs and memory consumption, hindering their application to process large-scale electronic health records (EHRs). We propose an efficient end-to-end model, the Joint-NER-RE-Fourier (JNRF), to jointly learn the tasks of named entity recognition and relation extraction for documents of variable length. The architecture uses positional encoding and unitary batch sizes to process variable length documents and uses a weight-shared Fourier network layer for low-complexity token mixing. Finally, we reach the theoretical computational complexity lower bound for relation extraction using a selective pooling strategy and distance-aware attention weights with trainable polynomial distance functions. We evaluated the JNRF architecture using the 2018 N2C2 ADE benchmark to jointly extract medication-related entities and relations in variable-length EHR summaries. JNRF outperforms rolling window BERT with selective pooling by 0.42%, while being twice as fast to train. Compared to state-of-the-art BiLSTM-CRF architectures on the N2C2 ADE benchmark, results show that the proposed approach trains 22 times faster and reduces GPU memory consumption by 1.75 folds, with a reasonable performance tradeoff of 90%, without the use of external tools, hand-crafted rules or post-processing. Given the significant carbon footprint of deep learning models and the current energy crises, these methods could support efficient and cleaner information extraction in EHRs and other types of large-scale document databases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge