Ihor Stepanov
GLiClass: Generalist Lightweight Model for Sequence Classification Tasks
Aug 11, 2025Abstract:Classification is one of the most widespread tasks in AI applications, serving often as the first step in filtering, sorting, and categorizing data. Since modern AI systems must handle large volumes of input data and early pipeline stages can propagate errors downstream, achieving high efficiency and accuracy is critical. Moreover, classification requirements can change dynamically based on user needs, necessitating models with strong zero-shot capabilities. While generative LLMs have become mainstream for zero-shot classification due to their versatility, they suffer from inconsistent instruction following and computational inefficiency. Cross-encoders, commonly used as rerankers in RAG pipelines, face a different bottleneck: they must process text-label pairs sequentially, significantly reducing efficiency with large label sets. Embedding-based approaches offer good efficiency but struggle with complex scenarios involving logical and semantic constraints. We propose GLiClass, a novel method that adapts the GLiNER architecture for sequence classification tasks. Our approach achieves strong accuracy and efficiency comparable to embedding-based methods, while maintaining the flexibility needed for zero-shot and few-shot learning scenarios. Additionally, we adapted proximal policy optimization (PPO) for multi-label text classification, enabling training classifiers in data-sparse conditions or from human feedback.
GLiNER-biomed: A Suite of Efficient Models for Open Biomedical Named Entity Recognition
Apr 01, 2025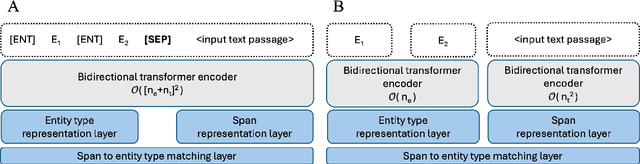



Abstract:Biomedical named entity recognition (NER) presents unique challenges due to specialized vocabularies, the sheer volume of entities, and the continuous emergence of novel entities. Traditional NER models, constrained by fixed taxonomies and human annotations, struggle to generalize beyond predefined entity types or efficiently adapt to emerging concepts. To address these issues, we introduce GLiNER-biomed, a domain-adapted suite of Generalist and Lightweight Model for NER (GLiNER) models specifically tailored for biomedical NER. In contrast to conventional approaches, GLiNER uses natural language descriptions to infer arbitrary entity types, enabling zero-shot recognition. Our approach first distills the annotation capabilities of large language models (LLMs) into a smaller, more efficient model, enabling the generation of high-coverage synthetic biomedical NER data. We subsequently train two GLiNER architectures, uni- and bi-encoder, at multiple scales to balance computational efficiency and recognition performance. Evaluations on several biomedical datasets demonstrate that GLiNER-biomed outperforms state-of-the-art GLiNER models in both zero- and few-shot scenarios, achieving 5.96% improvement in F1-score over the strongest baseline. Ablation studies highlight the effectiveness of our synthetic data generation strategy and emphasize the complementary benefits of synthetic biomedical pre-training combined with fine-tuning on high-quality general-domain annotations. All datasets, models, and training pipelines are publicly available at https://github.com/ds4dh/GLiNER-biomed.
GLiNER multi-task: Generalist Lightweight Model for Various Information Extraction Tasks
Jun 14, 2024Abstract:Information extraction tasks require both accurate, efficient, and generalisable models. Classical supervised deep learning approaches can achieve the required performance, but they need large datasets and are limited in their ability to adapt to different tasks. On the other hand, large language models (LLMs) demonstrate good generalization, meaning that they can adapt to many different tasks based on user requests. However, LLMs are computationally expensive and tend to fail to generate structured outputs. In this article, we will introduce a new kind of GLiNER model that can be used for various information extraction tasks while being a small encoder model. Our model achieved SoTA performance on zero-shot NER benchmarks and leading performance on question-answering, summarization and relation extraction tasks. Additionally, in this article, we will cover experimental results on self-learning approaches for named entity recognition using GLiNER models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge