Haiteng Zhao
Long-horizon Reasoning Agent for Olympiad-Level Mathematical Problem Solving
Dec 12, 2025Abstract:Large Reasoning Models (LRMs) have expanded the mathematical reasoning frontier through Chain-of-Thought (CoT) techniques and Reinforcement Learning with Verifiable Rewards (RLVR), capable of solving AIME-level problems. However, the performance of LRMs is heavily dependent on the extended reasoning context length. For solving ultra-hard problems like those in the International Mathematical Olympiad (IMO), the required reasoning complexity surpasses the space that an LRM can explore in a single round. Previous works attempt to extend the reasoning context of LRMs but remain prompt-based and built upon proprietary models, lacking systematic structures and training pipelines. Therefore, this paper introduces Intern-S1-MO, a long-horizon math agent that conducts multi-round hierarchical reasoning, composed of an LRM-based multi-agent system including reasoning, summary, and verification. By maintaining a compact memory in the form of lemmas, Intern-S1-MO can more freely explore the lemma-rich reasoning spaces in multiple reasoning stages, thereby breaking through the context constraints for IMO-level math problems. Furthermore, we propose OREAL-H, an RL framework for training the LRM using the online explored trajectories to simultaneously bootstrap the reasoning ability of LRM and elevate the overall performance of Intern-S1-MO. Experiments show that Intern-S1-MO can obtain 26 out of 35 points on the non-geometry problems of IMO2025, matching the performance of silver medalists. It also surpasses the current advanced LRMs on inference benchmarks such as HMMT2025, AIME2025, and CNMO2025. In addition, our agent officially participates in CMO2025 and achieves a score of 102/126 under the judgment of human experts, reaching the gold medal level.
Achieving Olympia-Level Geometry Large Language Model Agent via Complexity Boosting Reinforcement Learning
Dec 12, 2025Abstract:Large language model (LLM) agents exhibit strong mathematical problem-solving abilities and can even solve International Mathematical Olympiad (IMO) level problems with the assistance of formal proof systems. However, due to weak heuristics for auxiliary constructions, AI for geometry problem solving remains dominated by expert models such as AlphaGeometry 2, which rely heavily on large-scale data synthesis and search for both training and evaluation. In this work, we make the first attempt to build a medalist-level LLM agent for geometry and present InternGeometry. InternGeometry overcomes the heuristic limitations in geometry by iteratively proposing propositions and auxiliary constructions, verifying them with a symbolic engine, and reflecting on the engine's feedback to guide subsequent proposals. A dynamic memory mechanism enables InternGeometry to conduct more than two hundred interactions with the symbolic engine per problem. To further accelerate learning, we introduce Complexity-Boosting Reinforcement Learning (CBRL), which gradually increases the complexity of synthesized problems across training stages. Built on InternThinker-32B, InternGeometry solves 44 of 50 IMO geometry problems (2000-2024), exceeding the average gold medalist score (40.9), using only 13K training examples, just 0.004% of the data used by AlphaGeometry 2, demonstrating the potential of LLM agents on expert-level geometry tasks. InternGeometry can also propose novel auxiliary constructions for IMO problems that do not appear in human solutions. We will release the model, data, and symbolic engine to support future research.
Semi-off-Policy Reinforcement Learning for Vision-Language Slow-thinking Reasoning
Jul 22, 2025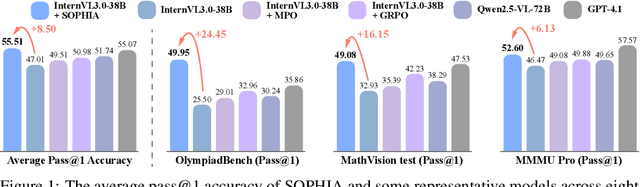
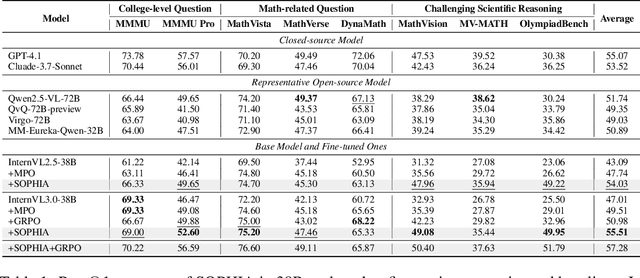
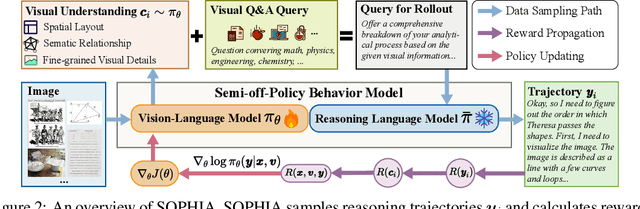
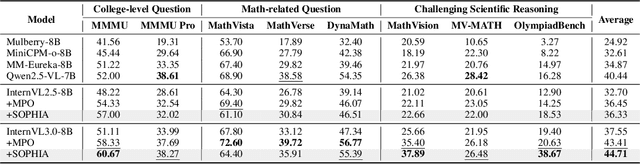
Abstract:Enhancing large vision-language models (LVLMs) with visual slow-thinking reasoning is crucial for solving complex multimodal tasks. However, since LVLMs are mainly trained with vision-language alignment, it is difficult to adopt on-policy reinforcement learning (RL) to develop the slow thinking ability because the rollout space is restricted by its initial abilities. Off-policy RL offers a way to go beyond the current policy, but directly distilling trajectories from external models may cause visual hallucinations due to mismatched visual perception abilities across models. To address these issues, this paper proposes SOPHIA, a simple and scalable Semi-Off-Policy RL for vision-language slow-tHInking reAsoning. SOPHIA builds a semi-off-policy behavior model by combining on-policy visual understanding from a trainable LVLM with off-policy slow-thinking reasoning from a language model, assigns outcome-based rewards to reasoning, and propagates visual rewards backward. Then LVLM learns slow-thinking reasoning ability from the obtained reasoning trajectories using propagated rewards via off-policy RL algorithms. Extensive experiments with InternVL2.5 and InternVL3.0 with 8B and 38B sizes show the effectiveness of SOPHIA. Notably, SOPHIA improves InternVL3.0-38B by 8.50% in average, reaching state-of-the-art performance among open-source LVLMs on multiple multimodal reasoning benchmarks, and even outperforms some closed-source models (e.g., GPT-4.1) on the challenging MathVision and OlympiadBench, achieving 49.08% and 49.95% pass@1 accuracy, respectively. Analysis shows SOPHIA outperforms supervised fine-tuning and direct on-policy RL methods, offering a better policy initialization for further on-policy training.
ScienceBoard: Evaluating Multimodal Autonomous Agents in Realistic Scientific Workflows
May 26, 2025



Abstract:Large Language Models (LLMs) have extended their impact beyond Natural Language Processing, substantially fostering the development of interdisciplinary research. Recently, various LLM-based agents have been developed to assist scientific discovery progress across multiple aspects and domains. Among these, computer-using agents, capable of interacting with operating systems as humans do, are paving the way to automated scientific problem-solving and addressing routines in researchers' workflows. Recognizing the transformative potential of these agents, we introduce ScienceBoard, which encompasses two complementary contributions: (i) a realistic, multi-domain environment featuring dynamic and visually rich scientific workflows with integrated professional software, where agents can autonomously interact via different interfaces to accelerate complex research tasks and experiments; and (ii) a challenging benchmark of 169 high-quality, rigorously validated real-world tasks curated by humans, spanning scientific-discovery workflows in domains such as biochemistry, astronomy, and geoinformatics. Extensive evaluations of agents with state-of-the-art backbones (e.g., GPT-4o, Claude 3.7, UI-TARS) show that, despite some promising results, they still fall short of reliably assisting scientists in complex workflows, achieving only a 15% overall success rate. In-depth analysis further provides valuable insights for addressing current agent limitations and more effective design principles, paving the way to build more capable agents for scientific discovery. Our code, environment, and benchmark are at https://qiushisun.github.io/ScienceBoard-Home/.
Genius: A Generalizable and Purely Unsupervised Self-Training Framework For Advanced Reasoning
Apr 11, 2025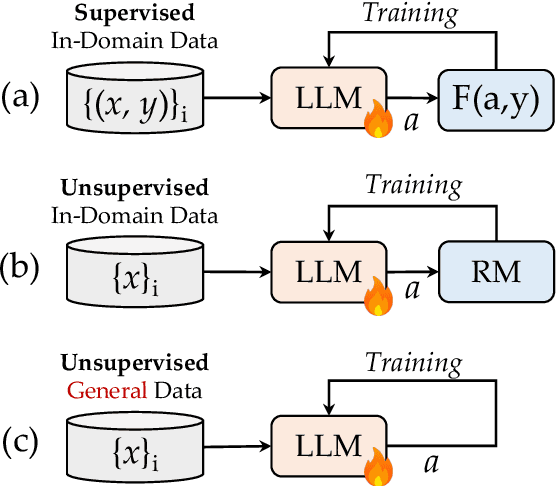
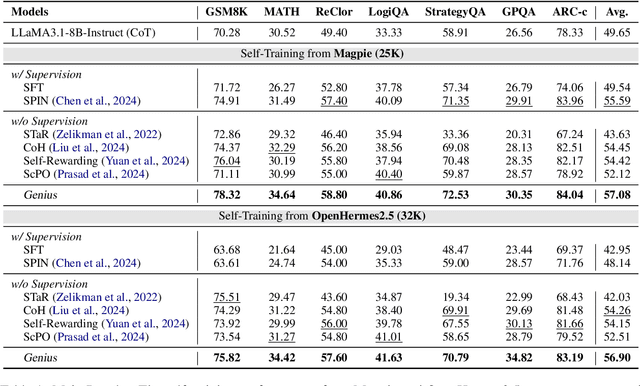
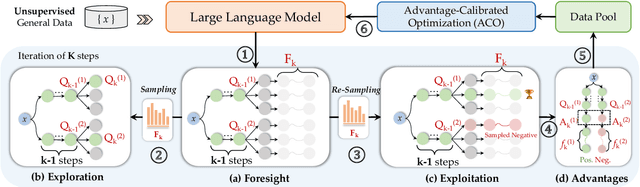
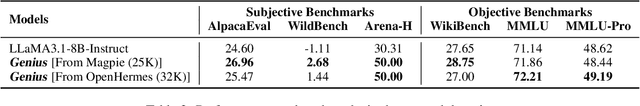
Abstract:Advancing LLM reasoning skills has captivated wide interest. However, current post-training techniques rely heavily on supervisory signals, such as outcome supervision or auxiliary reward models, which face the problem of scalability and high annotation costs. This motivates us to enhance LLM reasoning without the need for external supervision. We introduce a generalizable and purely unsupervised self-training framework, named Genius. Without external auxiliary, Genius requires to seek the optimal response sequence in a stepwise manner and optimize the LLM. To explore the potential steps and exploit the optimal ones, Genius introduces a stepwise foresight re-sampling strategy to sample and estimate the step value by simulating future outcomes. Further, we recognize that the unsupervised setting inevitably induces the intrinsic noise and uncertainty. To provide a robust optimization, we propose an advantage-calibrated optimization (ACO) loss function to mitigate estimation inconsistencies. Combining these techniques together, Genius provides an advanced initial step towards self-improve LLM reasoning with general queries and without supervision, revolutionizing reasoning scaling laws given the vast availability of general queries. The code will be released at https://github.com/xufangzhi/Genius.
$φ$-Decoding: Adaptive Foresight Sampling for Balanced Inference-Time Exploration and Exploitation
Mar 17, 2025Abstract:Inference-time optimization scales computation to derive deliberate reasoning steps for effective performance. While previous search-based strategies address the short-sightedness of auto-regressive generation, the vast search space leads to excessive exploration and insufficient exploitation. To strike an efficient balance to derive the optimal step, we frame the decoding strategy as foresight sampling, leveraging simulated future steps to obtain globally optimal step estimation. Built on it, we propose a novel decoding strategy, named $\phi$-Decoding. To provide a precise and expressive estimation of step value, $\phi$-Decoding approximates two distributions via foresight and clustering. Sampling from the joint distribution, the optimal steps can be selected for exploitation. To support adaptive computation allocation, we propose in-width and in-depth pruning strategies, featuring a light-weight solution to achieve inference efficiency. Extensive experiments across seven benchmarks show $\phi$-Decoding outperforms strong baselines in both performance and efficiency. Additional analysis demonstrates its generalization across various LLMs and scalability across a wide range of computing budgets. The code will be released at https://github.com/xufangzhi/phi-Decoding, and the open-source PyPI package is coming soon.
BioMaze: Benchmarking and Enhancing Large Language Models for Biological Pathway Reasoning
Feb 23, 2025Abstract:The applications of large language models (LLMs) in various biological domains have been explored recently, but their reasoning ability in complex biological systems, such as pathways, remains underexplored, which is crucial for predicting biological phenomena, formulating hypotheses, and designing experiments. This work explores the potential of LLMs in pathway reasoning. We introduce BioMaze, a dataset with 5.1K complex pathway problems derived from real research, covering various biological contexts including natural dynamic changes, disturbances, additional intervention conditions, and multi-scale research targets. Our evaluation of methods such as CoT and graph-augmented reasoning, shows that LLMs struggle with pathway reasoning, especially in perturbed systems. To address this, we propose PathSeeker, an LLM agent that enhances reasoning through interactive subgraph-based navigation, enabling a more effective approach to handling the complexities of biological systems in a scientifically aligned manner. The dataset and code are available at https://github.com/zhao-ht/BioMaze.
Non-myopic Generation of Language Models for Reasoning and Planning
Oct 28, 2024



Abstract:Large Language Models have demonstrated remarkable abilities in reasoning and planning by breaking down complex problems into sequential steps. Despite their success in various domains like mathematical problem-solving and coding, LLMs face challenges in ensuring reliable and optimal planning due to their inherent myopic nature of autoregressive decoding. This paper revisits LLM reasoning from an optimal-control perspective, proposing a novel method, Predictive-Decoding, that leverages Model Predictive Control to enhance planning accuracy. By re-weighting LLM distributions based on foresight trajectories, Predictive-Decoding aims to mitigate early errors and promote non-myopic planning. Our experiments show significant improvements in a wide range of tasks for math, coding, and agents. Furthermore, Predictive-Decoding demonstrates computational efficiency, outperforming search baselines with reduced computational resources. This study provides insights into optimizing LLM planning capabilities.
Non-myopic Generation of Language Model for Reasoning and Planning
Oct 23, 2024



Abstract:Large Language Models have demonstrated remarkable abilities in reasoning and planning by breaking down complex problems into sequential steps. Despite their success in various domains like mathematical problem-solving and coding, LLMs face challenges in ensuring reliable and optimal planning due to their inherent myopic nature of autoregressive decoding. This paper revisits LLM reasoning from an optimal-control perspective, proposing a novel method, Predictive-Decoding, that leverages Model Predictive Control to enhance planning accuracy. By re-weighting LLM distributions based on foresight trajectories, Predictive-Decoding aims to mitigate early errors and promote non-myopic planning. Our experiments show significant improvements in a wide range of tasks for math, coding, and agents. Furthermore, Predictive-Decoding demonstrates computational efficiency, outperforming search baselines with reduced computational resources. This study provides insights into optimizing LLM planning capabilities.
Instruction-Based Molecular Graph Generation with Unified Text-Graph Diffusion Model
Aug 19, 2024Abstract:Recent advancements in computational chemistry have increasingly focused on synthesizing molecules based on textual instructions. Integrating graph generation with these instructions is complex, leading most current methods to use molecular sequences with pre-trained large language models. In response to this challenge, we propose a novel framework, named $\textbf{UTGDiff (Unified Text-Graph Diffusion Model)}$, which utilizes language models for discrete graph diffusion to generate molecular graphs from instructions. UTGDiff features a unified text-graph transformer as the denoising network, derived from pre-trained language models and minimally modified to process graph data through attention bias. Our experimental results demonstrate that UTGDiff consistently outperforms sequence-based baselines in tasks involving instruction-based molecule generation and editing, achieving superior performance with fewer parameters given an equivalent level of pretraining corpus. Our code is availble at https://github.com/ran1812/UTGDiff.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge