Dingshuo Chen
Materials Generation in the Era of Artificial Intelligence: A Comprehensive Survey
May 22, 2025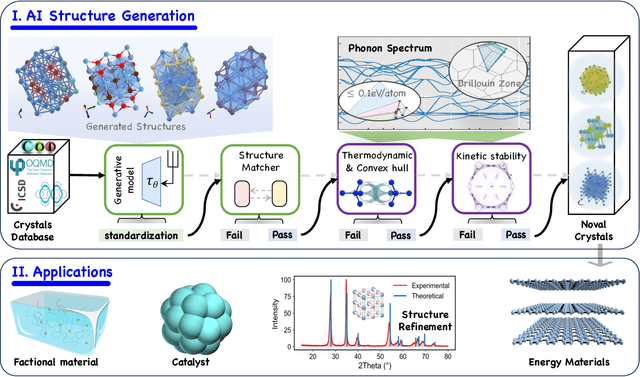
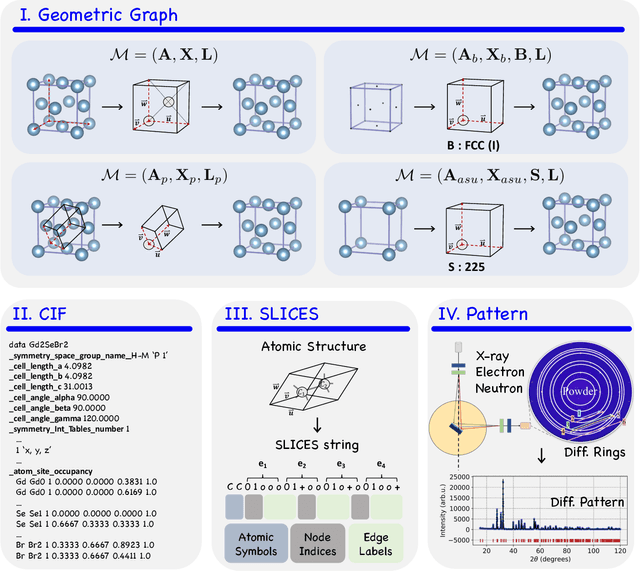
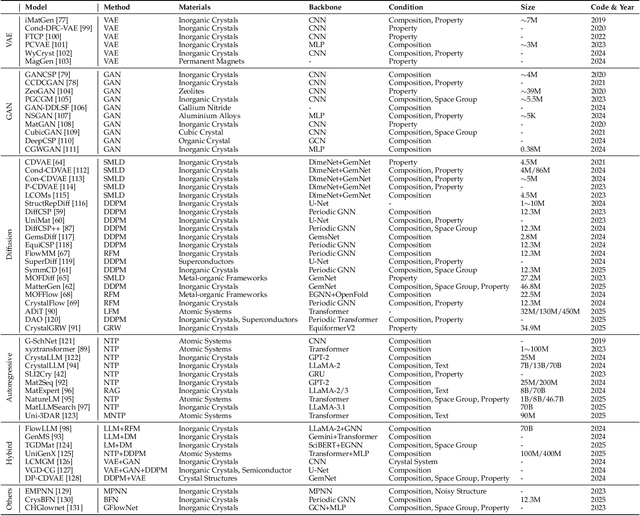
Abstract:Materials are the foundation of modern society, underpinning advancements in energy, electronics, healthcare, transportation, and infrastructure. The ability to discover and design new materials with tailored properties is critical to solving some of the most pressing global challenges. In recent years, the growing availability of high-quality materials data combined with rapid advances in Artificial Intelligence (AI) has opened new opportunities for accelerating materials discovery. Data-driven generative models provide a powerful tool for materials design by directly create novel materials that satisfy predefined property requirements. Despite the proliferation of related work, there remains a notable lack of up-to-date and systematic surveys in this area. To fill this gap, this paper provides a comprehensive overview of recent progress in AI-driven materials generation. We first organize various types of materials and illustrate multiple representations of crystalline materials. We then provide a detailed summary and taxonomy of current AI-driven materials generation approaches. Furthermore, we discuss the common evaluation metrics and summarize open-source codes and benchmark datasets. Finally, we conclude with potential future directions and challenges in this fast-growing field. The related sources can be found at https://github.com/ZhixunLEE/Awesome-AI-for-Materials-Generation.
Graffe: Graph Representation Learning via Diffusion Probabilistic Models
May 08, 2025Abstract:Diffusion probabilistic models (DPMs), widely recognized for their potential to generate high-quality samples, tend to go unnoticed in representation learning. While recent progress has highlighted their potential for capturing visual semantics, adapting DPMs to graph representation learning remains in its infancy. In this paper, we introduce Graffe, a self-supervised diffusion model proposed for graph representation learning. It features a graph encoder that distills a source graph into a compact representation, which, in turn, serves as the condition to guide the denoising process of the diffusion decoder. To evaluate the effectiveness of our model, we first explore the theoretical foundations of applying diffusion models to representation learning, proving that the denoising objective implicitly maximizes the conditional mutual information between data and its representation. Specifically, we prove that the negative logarithm of the denoising score matching loss is a tractable lower bound for the conditional mutual information. Empirically, we conduct a series of case studies to validate our theoretical insights. In addition, Graffe delivers competitive results under the linear probing setting on node and graph classification tasks, achieving state-of-the-art performance on 9 of the 11 real-world datasets. These findings indicate that powerful generative models, especially diffusion models, serve as an effective tool for graph representation learning.
IceBerg: Debiased Self-Training for Class-Imbalanced Node Classification
Feb 10, 2025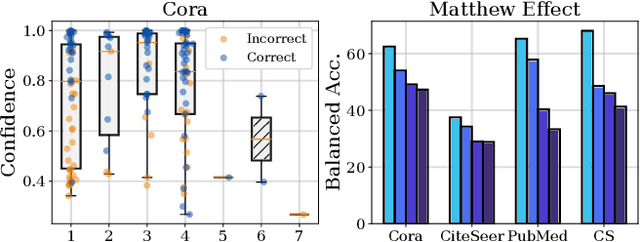
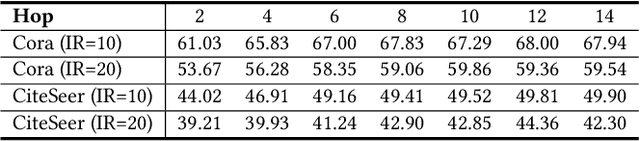
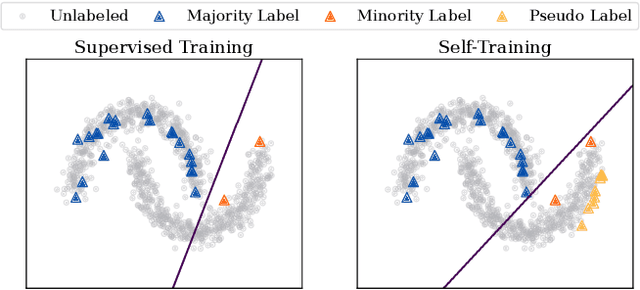
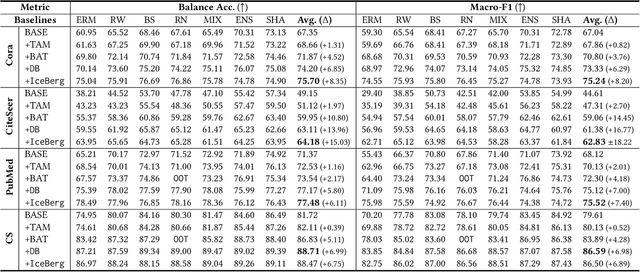
Abstract:Graph Neural Networks (GNNs) have achieved great success in dealing with non-Euclidean graph-structured data and have been widely deployed in many real-world applications. However, their effectiveness is often jeopardized under class-imbalanced training sets. Most existing studies have analyzed class-imbalanced node classification from a supervised learning perspective, but they do not fully utilize the large number of unlabeled nodes in semi-supervised scenarios. We claim that the supervised signal is just the tip of the iceberg and a large number of unlabeled nodes have not yet been effectively utilized. In this work, we propose IceBerg, a debiased self-training framework to address the class-imbalanced and few-shot challenges for GNNs at the same time. Specifically, to figure out the Matthew effect and label distribution shift in self-training, we propose Double Balancing, which can largely improve the performance of existing baselines with just a few lines of code as a simple plug-and-play module. Secondly, to enhance the long-range propagation capability of GNNs, we disentangle the propagation and transformation operations of GNNs. Therefore, the weak supervision signals can propagate more effectively to address the few-shot issue. In summary, we find that leveraging unlabeled nodes can significantly enhance the performance of GNNs in class-imbalanced and few-shot scenarios, and even small, surgical modifications can lead to substantial performance improvements. Systematic experiments on benchmark datasets show that our method can deliver considerable performance gain over existing class-imbalanced node classification baselines. Additionally, due to IceBerg's outstanding ability to leverage unsupervised signals, it also achieves state-of-the-art results in few-shot node classification scenarios. The code of IceBerg is available at: https://github.com/ZhixunLEE/IceBerg.
GDeR: Safeguarding Efficiency, Balancing, and Robustness via Prototypical Graph Pruning
Oct 17, 2024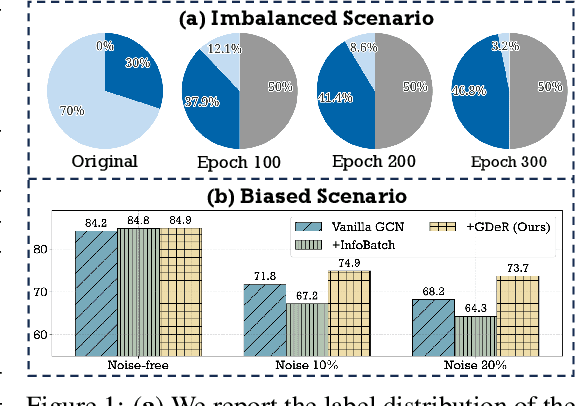
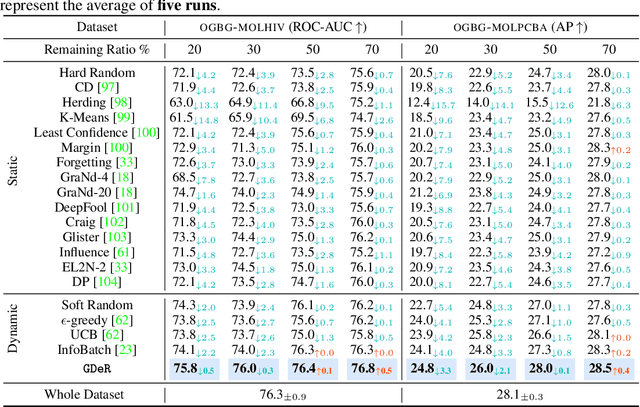
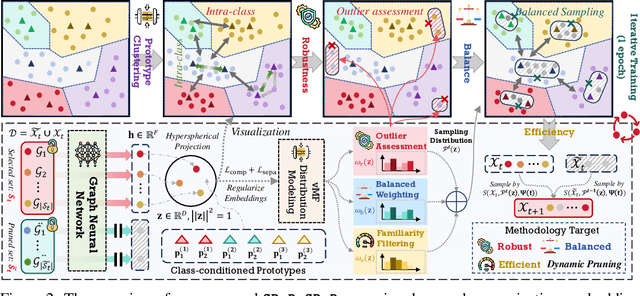
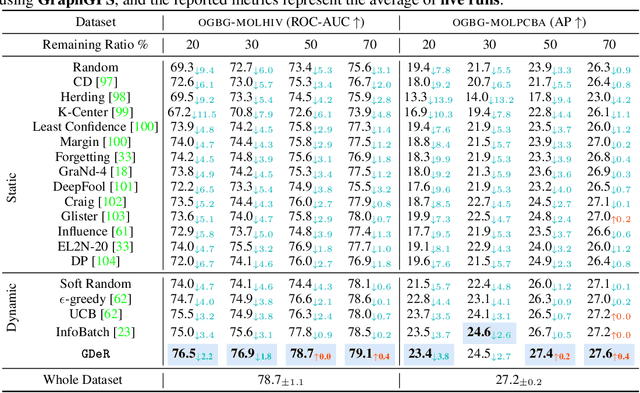
Abstract:Training high-quality deep models necessitates vast amounts of data, resulting in overwhelming computational and memory demands. Recently, data pruning, distillation, and coreset selection have been developed to streamline data volume by retaining, synthesizing, or selecting a small yet informative subset from the full set. Among these methods, data pruning incurs the least additional training cost and offers the most practical acceleration benefits. However, it is the most vulnerable, often suffering significant performance degradation with imbalanced or biased data schema, thus raising concerns about its accuracy and reliability in on-device deployment. Therefore, there is a looming need for a new data pruning paradigm that maintains the efficiency of previous practices while ensuring balance and robustness. Unlike the fields of computer vision and natural language processing, where mature solutions have been developed to address these issues, graph neural networks (GNNs) continue to struggle with increasingly large-scale, imbalanced, and noisy datasets, lacking a unified dataset pruning solution. To achieve this, we introduce a novel dynamic soft-pruning method, GDeR, designed to update the training ``basket'' during the process using trainable prototypes. GDeR first constructs a well-modeled graph embedding hypersphere and then samples \textit{representative, balanced, and unbiased subsets} from this embedding space, which achieves the goal we called Graph Training Debugging. Extensive experiments on five datasets across three GNN backbones, demonstrate that GDeR (I) achieves or surpasses the performance of the full dataset with 30%~50% fewer training samples, (II) attains up to a 2.81x lossless training speedup, and (III) outperforms state-of-the-art pruning methods in imbalanced training and noisy training scenarios by 0.3%~4.3% and 3.6%~7.8%, respectively.
Beyond Efficiency: Molecular Data Pruning for Enhanced Generalization
Sep 02, 2024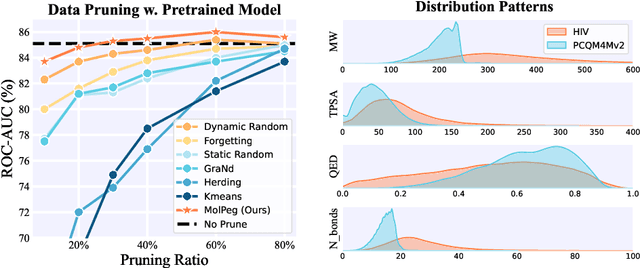
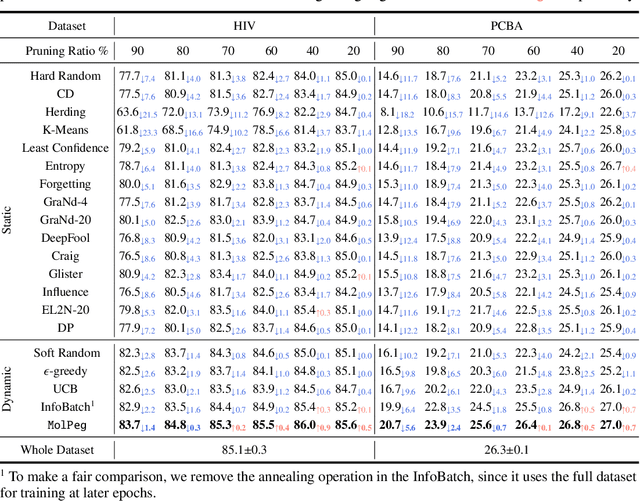
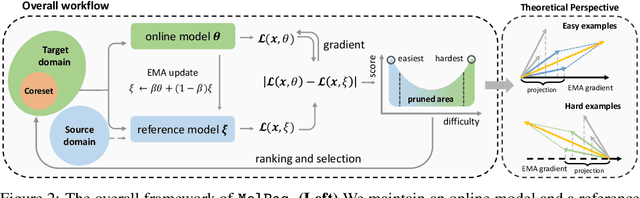

Abstract:With the emergence of various molecular tasks and massive datasets, how to perform efficient training has become an urgent yet under-explored issue in the area. Data pruning (DP), as an oft-stated approach to saving training burdens, filters out less influential samples to form a coreset for training. However, the increasing reliance on pretrained models for molecular tasks renders traditional in-domain DP methods incompatible. Therefore, we propose a Molecular data Pruning framework for enhanced Generalization (MolPeg), which focuses on the source-free data pruning scenario, where data pruning is applied with pretrained models. By maintaining two models with different updating paces during training, we introduce a novel scoring function to measure the informativeness of samples based on the loss discrepancy. As a plug-and-play framework, MolPeg realizes the perception of both source and target domain and consistently outperforms existing DP methods across four downstream tasks. Remarkably, it can surpass the performance obtained from full-dataset training, even when pruning up to 60-70% of the data on HIV and PCBA dataset. Our work suggests that the discovery of effective data-pruning metrics could provide a viable path to both enhanced efficiency and superior generalization in transfer learning.
GSLB: The Graph Structure Learning Benchmark
Oct 08, 2023Abstract:Graph Structure Learning (GSL) has recently garnered considerable attention due to its ability to optimize both the parameters of Graph Neural Networks (GNNs) and the computation graph structure simultaneously. Despite the proliferation of GSL methods developed in recent years, there is no standard experimental setting or fair comparison for performance evaluation, which creates a great obstacle to understanding the progress in this field. To fill this gap, we systematically analyze the performance of GSL in different scenarios and develop a comprehensive Graph Structure Learning Benchmark (GSLB) curated from 20 diverse graph datasets and 16 distinct GSL algorithms. Specifically, GSLB systematically investigates the characteristics of GSL in terms of three dimensions: effectiveness, robustness, and complexity. We comprehensively evaluate state-of-the-art GSL algorithms in node- and graph-level tasks, and analyze their performance in robust learning and model complexity. Further, to facilitate reproducible research, we have developed an easy-to-use library for training, evaluating, and visualizing different GSL methods. Empirical results of our extensive experiments demonstrate the ability of GSL and reveal its potential benefits on various downstream tasks, offering insights and opportunities for future research. The code of GSLB is available at: https://github.com/GSL-Benchmark/GSLB.
Uncovering Neural Scaling Laws in Molecular Representation Learning
Sep 28, 2023



Abstract:Molecular Representation Learning (MRL) has emerged as a powerful tool for drug and materials discovery in a variety of tasks such as virtual screening and inverse design. While there has been a surge of interest in advancing model-centric techniques, the influence of both data quantity and quality on molecular representations is not yet clearly understood within this field. In this paper, we delve into the neural scaling behaviors of MRL from a data-centric viewpoint, examining four key dimensions: (1) data modalities, (2) dataset splitting, (3) the role of pre-training, and (4) model capacity. Our empirical studies confirm a consistent power-law relationship between data volume and MRL performance across these dimensions. Additionally, through detailed analysis, we identify potential avenues for improving learning efficiency. To challenge these scaling laws, we adapt seven popular data pruning strategies to molecular data and benchmark their performance. Our findings underline the importance of data-centric MRL and highlight possible directions for future research.
Improving Molecular Pretraining with Complementary Featurizations
Sep 29, 2022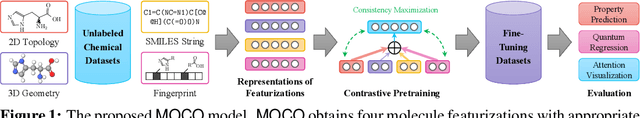
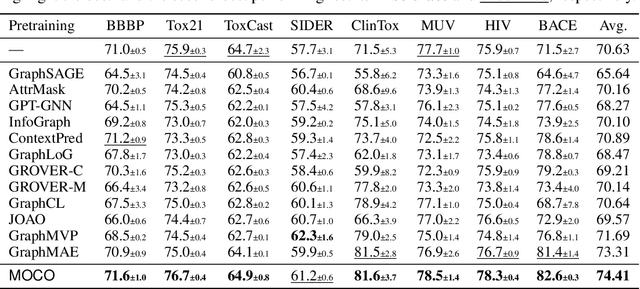
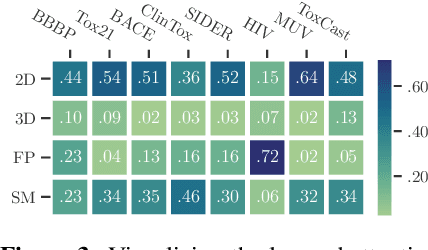
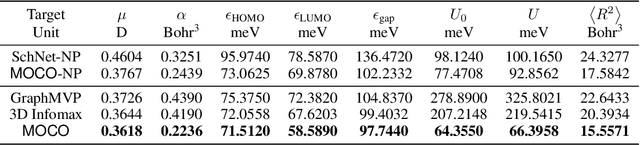
Abstract:Molecular pretraining, which learns molecular representations over massive unlabeled data, has become a prominent paradigm to solve a variety of tasks in computational chemistry and drug discovery. Recently, prosperous progress has been made in molecular pretraining with different molecular featurizations, including 1D SMILES strings, 2D graphs, and 3D geometries. However, the role of molecular featurizations with their corresponding neural architectures in molecular pretraining remains largely unexamined. In this paper, through two case studies -- chirality classification and aromatic ring counting -- we first demonstrate that different featurization techniques convey chemical information differently. In light of this observation, we propose a simple and effective MOlecular pretraining framework with COmplementary featurizations (MOCO). MOCO comprehensively leverages multiple featurizations that complement each other and outperforms existing state-of-the-art models that solely relies on one or two featurizations on a wide range of molecular property prediction tasks.
Learning Graph Structures with Transformer for Multivariate Time Series Anomaly Detection in IoT
Apr 08, 2021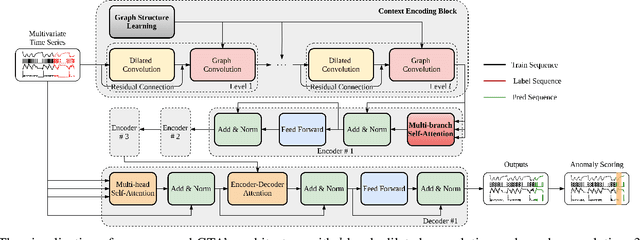
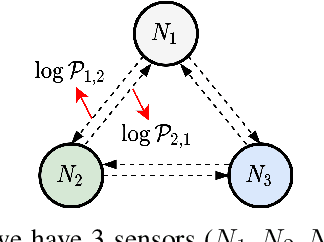
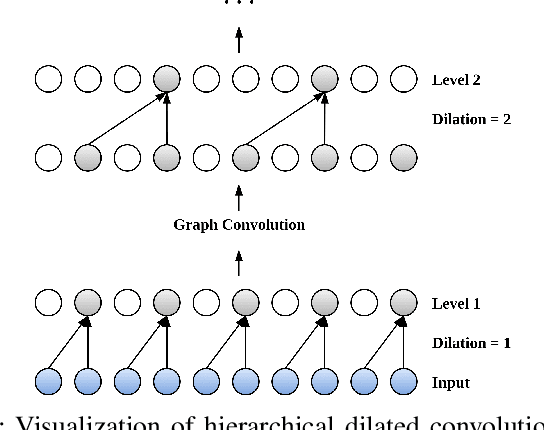
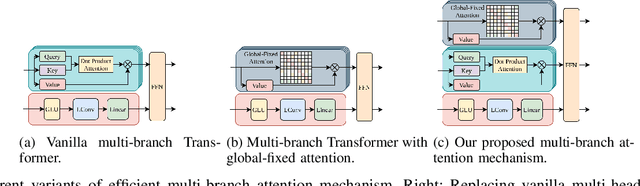
Abstract:Many real-world IoT systems comprising various internet-connected sensory devices generate substantial amounts of multivariate time series data. Meanwhile, those critical IoT infrastructures, such as smart power grids and water distribution networks, are often targets of cyber-attacks, making anomaly detection of high research value. However, considering the complex topological and nonlinear dependencies that are initially unknown among sensors, modeling such relatedness is inevitable for any efficient and accurate anomaly detection system. Additionally, due to multivariate time series' temporal dependency and stochasticity, their anomaly detection remains a big challenge. This work proposed a novel framework, namely GTA, for multivariate time series anomaly detection by automatically learning a graph structure followed by the graph convolution and modeling the temporal dependency through a Transformer-based architecture. The core idea of learning graph structure is called the connection learning policy based on the Gumbel-softmax sampling strategy to learn bi-directed associations among sensors directly. We also devised a novel graph convolution named Influence Propagation convolution to model the anomaly information flow between graph nodes. Moreover, we proposed a multi-branch attention mechanism to substitute for original multi-head self-attention to overcome the quadratic complexity challenge. The extensive experiments on four public anomaly detection benchmarks further demonstrate our approach's superiority over other state-of-the-arts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge