Ziqiang Xu
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Beyond English: Toward Inclusive and Scalable Multilingual Machine Translation with LLMs
Nov 10, 2025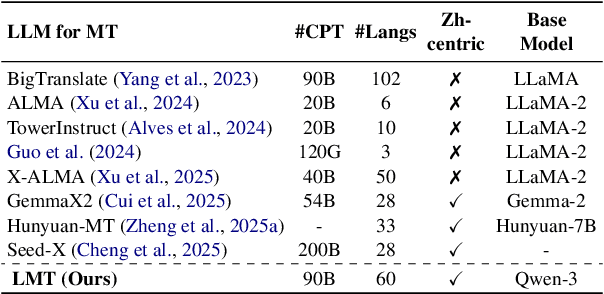
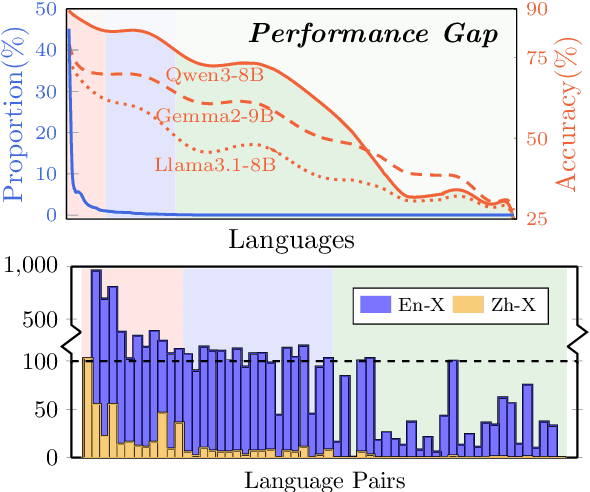
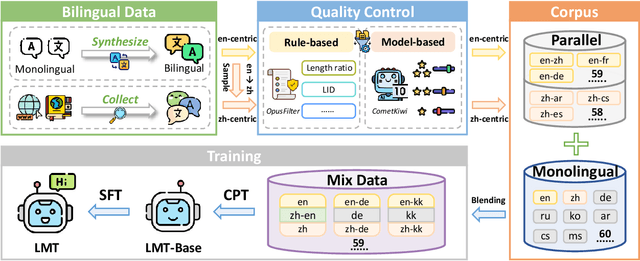
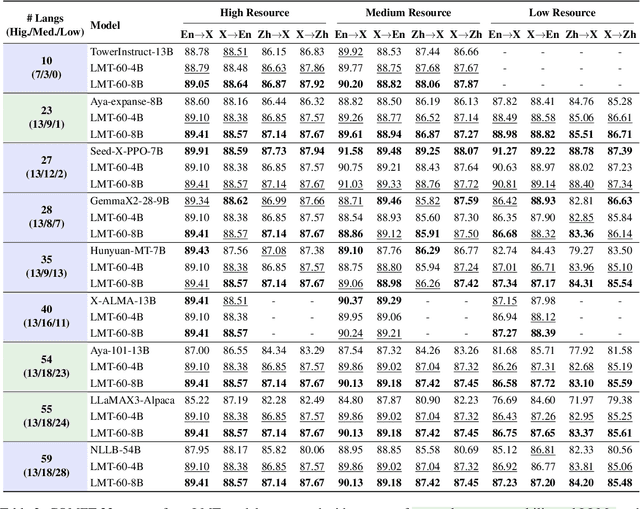
Abstract:Large language models have significantly advanced Multilingual Machine Translation (MMT), yet the broad language coverage, consistent translation quality, and English-centric bias remain open challenges. To address these challenges, we introduce \textbf{LMT}, a suite of \textbf{L}arge-scale \textbf{M}ultilingual \textbf{T}ranslation models centered on both Chinese and English, covering 60 languages and 234 translation directions. During development, we identify a previously overlooked phenomenon of \textbf{directional degeneration}, where symmetric multi-way fine-tuning data overemphasize reverse directions (X $\to$ En/Zh), leading to excessive many-to-one mappings and degraded translation quality. We propose \textbf{Strategic Downsampling}, a simple yet effective method to mitigate this degeneration. In addition, we design \textbf{Parallel Multilingual Prompting (PMP)}, which leverages typologically related auxiliary languages to enhance cross-lingual transfer. Through rigorous data curation and refined adaptation strategies, LMT achieves SOTA performance among models of comparable language coverage, with our 4B model (LMT-60-4B) surpassing the much larger Aya-101-13B and NLLB-54B models by a substantial margin. We release LMT in four sizes (0.6B/1.7B/4B/8B) to catalyze future research and provide strong baselines for inclusive, scalable, and high-quality MMT \footnote{\href{https://github.com/NiuTrans/LMT}{https://github.com/NiuTrans/LMT}}.
One Size Does Not Fit All: A Distribution-Aware Sparsification for More Precise Model Merging
Aug 08, 2025Abstract:Model merging has emerged as a compelling data-free paradigm for multi-task learning, enabling the fusion of multiple fine-tuned models into a single, powerful entity. A key technique in merging methods is sparsification, which prunes redundant parameters from task vectors to mitigate interference. However, prevailing approaches employ a ``one-size-fits-all'' strategy, applying a uniform sparsity ratio that overlooks the inherent structural and statistical heterogeneity of model parameters. This often leads to a suboptimal trade-off, where critical parameters are inadvertently pruned while less useful ones are retained. To address this limitation, we introduce \textbf{TADrop} (\textbf{T}ensor-wise \textbf{A}daptive \textbf{Drop}), an adaptive sparsification strategy that respects this heterogeneity. Instead of a global ratio, TADrop assigns a tailored sparsity level to each parameter tensor based on its distributional properties. The core intuition is that tensors with denser, more redundant distributions can be pruned aggressively, while sparser, more critical ones are preserved. As a simple and plug-and-play module, we validate TADrop by integrating it with foundational, classic, and SOTA merging methods. Extensive experiments across diverse tasks (vision, language, and multimodal) and models (ViT, BEiT) demonstrate that TADrop consistently and significantly boosts their performance. For instance, when enhancing a leading merging method, it achieves an average performance gain of 2.0\% across 8 ViT-B/32 tasks. TADrop provides a more effective way to mitigate parameter interference by tailoring sparsification to the model's structure, offering a new baseline for high-performance model merging.
Phi-Ground Tech Report: Advancing Perception in GUI Grounding
Jul 31, 2025



Abstract:With the development of multimodal reasoning models, Computer Use Agents (CUAs), akin to Jarvis from \textit{"Iron Man"}, are becoming a reality. GUI grounding is a core component for CUAs to execute actual actions, similar to mechanical control in robotics, and it directly leads to the success or failure of the system. It determines actions such as clicking and typing, as well as related parameters like the coordinates for clicks. Current end-to-end grounding models still achieve less than 65\% accuracy on challenging benchmarks like ScreenSpot-pro and UI-Vision, indicating they are far from being ready for deployment. % , as a single misclick can result in unacceptable consequences. In this work, we conduct an empirical study on the training of grounding models, examining details from data collection to model training. Ultimately, we developed the \textbf{Phi-Ground} model family, which achieves state-of-the-art performance across all five grounding benchmarks for models under $10B$ parameters in agent settings. In the end-to-end model setting, our model still achieves SOTA results with scores of \textit{\textbf{43.2}} on ScreenSpot-pro and \textit{\textbf{27.2}} on UI-Vision. We believe that the various details discussed in this paper, along with our successes and failures, not only clarify the construction of grounding models but also benefit other perception tasks. Project homepage: \href{https://zhangmiaosen2000.github.io/Phi-Ground/}{https://zhangmiaosen2000.github.io/Phi-Ground/}
ViaRL: Adaptive Temporal Grounding via Visual Iterated Amplification Reinforcement Learning
May 21, 2025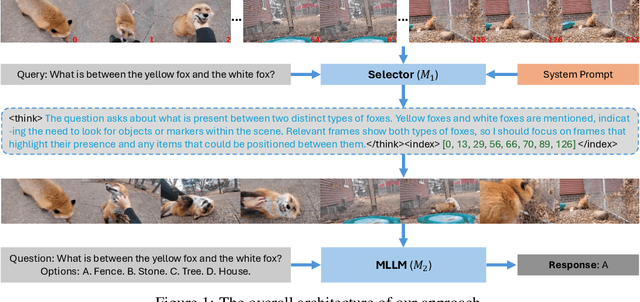
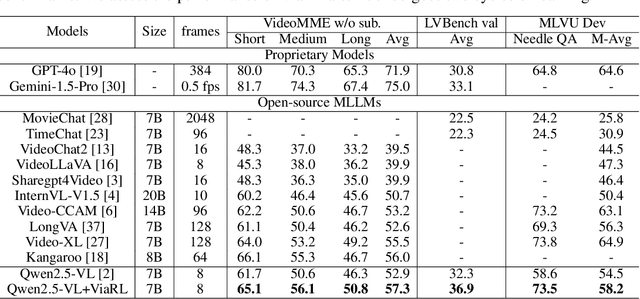
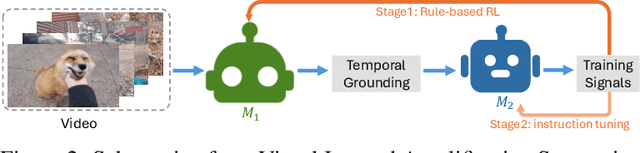
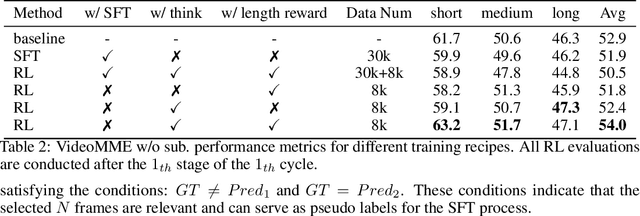
Abstract:Video understanding is inherently intention-driven-humans naturally focus on relevant frames based on their goals. Recent advancements in multimodal large language models (MLLMs) have enabled flexible query-driven reasoning; however, video-based frameworks like Video Chain-of-Thought lack direct training signals to effectively identify relevant frames. Current approaches often rely on heuristic methods or pseudo-label supervised annotations, which are both costly and limited in scalability across diverse scenarios. To overcome these challenges, we introduce ViaRL, the first framework to leverage rule-based reinforcement learning (RL) for optimizing frame selection in intention-driven video understanding. An iterated amplification strategy is adopted to perform alternating cyclic training in the video CoT system, where each component undergoes iterative cycles of refinement to improve its capabilities. ViaRL utilizes the answer accuracy of a downstream model as a reward signal to train a frame selector through trial-and-error, eliminating the need for expensive annotations while closely aligning with human-like learning processes. Comprehensive experiments across multiple benchmarks, including VideoMME, LVBench, and MLVU, demonstrate that ViaRL consistently delivers superior temporal grounding performance and robust generalization across diverse video understanding tasks, highlighting its effectiveness and scalability. Notably, ViaRL achieves a nearly 15\% improvement on Needle QA, a subset of MLVU, which is required to search a specific needle within a long video and regarded as one of the most suitable benchmarks for evaluating temporal grounding.
Beyond Decoder-only: Large Language Models Can be Good Encoders for Machine Translation
Mar 09, 2025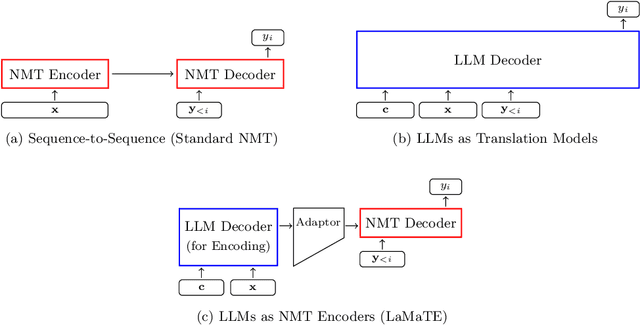
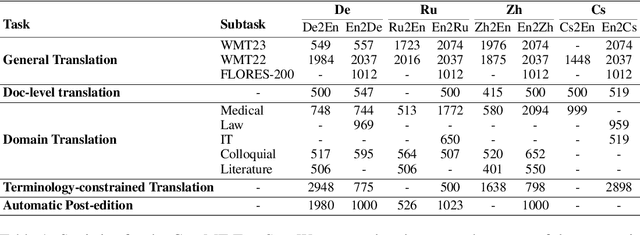
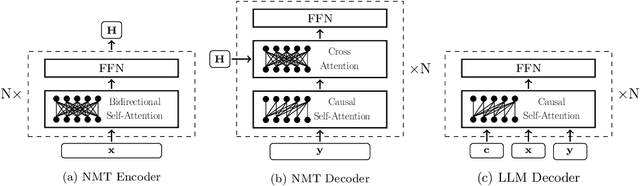
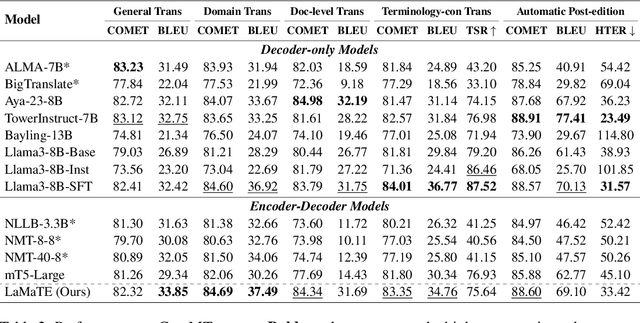
Abstract:The field of neural machine translation (NMT) has changed with the advent of large language models (LLMs). Much of the recent emphasis in natural language processing (NLP) has been on modeling machine translation and many other problems using a single pre-trained Transformer decoder, while encoder-decoder architectures, which were the standard in earlier NMT models, have received relatively less attention. In this paper, we explore translation models that are universal, efficient, and easy to optimize, by marrying the world of LLMs with the world of NMT. We apply LLMs to NMT encoding and leave the NMT decoder unchanged. We also develop methods for adapting LLMs to work better with the NMT decoder. Furthermore, we construct a new dataset involving multiple tasks to assess how well the machine translation system generalizes across various tasks. Evaluations on the WMT and our datasets show that results using our method match or surpass a range of baselines in terms of translation quality, but achieve $2.4 \sim 6.5 \times$ inference speedups and a $75\%$ reduction in the memory footprint of the KV cache. It also demonstrates strong generalization across a variety of translation-related tasks.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
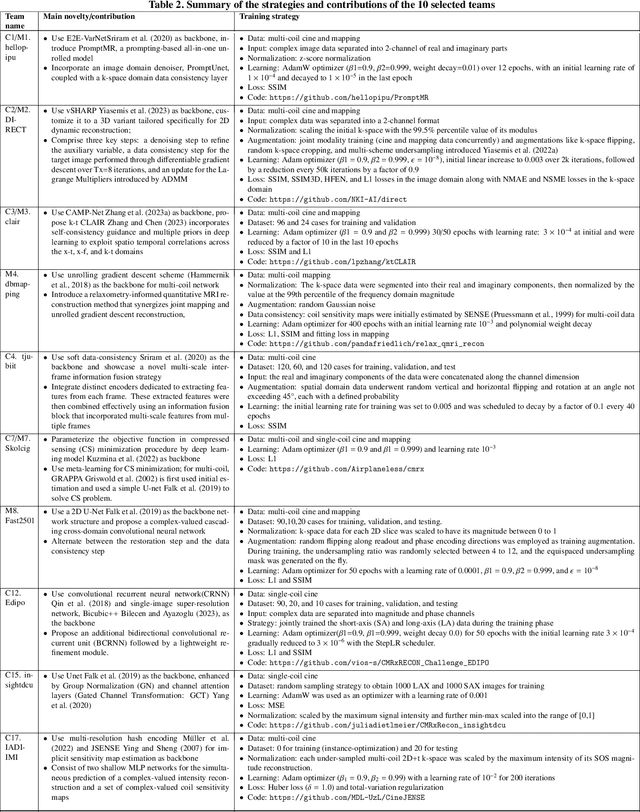
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
Multi-task SAR Image Processing via GAN-based Unsupervised Manipulation
Aug 02, 2024



Abstract:Generative Adversarial Networks (GANs) have shown tremendous potential in synthesizing a large number of realistic SAR images by learning patterns in the data distribution. Some GANs can achieve image editing by introducing latent codes, demonstrating significant promise in SAR image processing. Compared to traditional SAR image processing methods, editing based on GAN latent space control is entirely unsupervised, allowing image processing to be conducted without any labeled data. Additionally, the information extracted from the data is more interpretable. This paper proposes a novel SAR image processing framework called GAN-based Unsupervised Editing (GUE), aiming to address the following two issues: (1) disentangling semantic directions in the GAN latent space and finding meaningful directions; (2) establishing a comprehensive SAR image processing framework while achieving multiple image processing functions. In the implementation of GUE, we decompose the entangled semantic directions in the GAN latent space by training a carefully designed network. Moreover, we can accomplish multiple SAR image processing tasks (including despeckling, localization, auxiliary identification, and rotation editing) in a single training process without any form of supervision. Extensive experiments validate the effectiveness of the proposed method.
CMRxRecon2024: A Multi-Modality, Multi-View K-Space Dataset Boosting Universal Machine Learning for Accelerated Cardiac MRI
Jun 27, 2024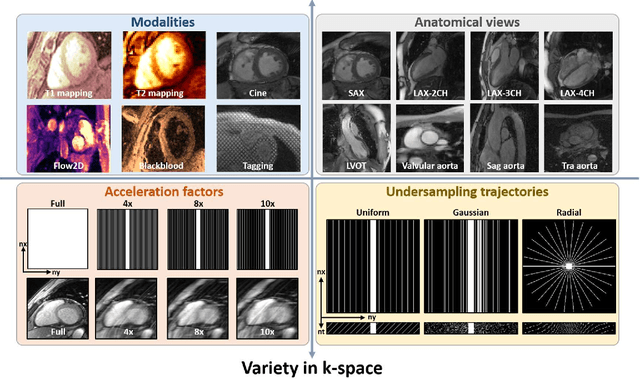
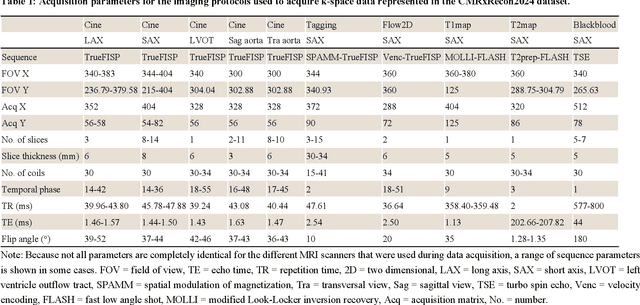
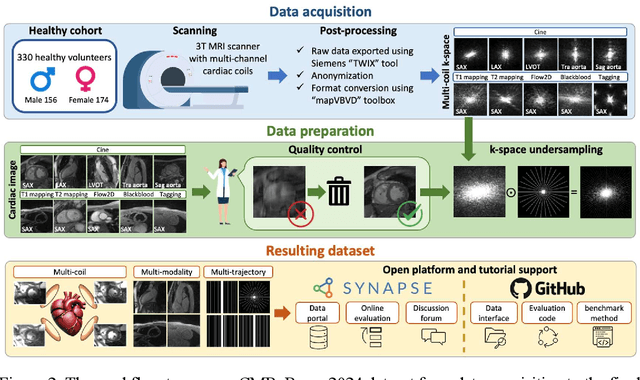
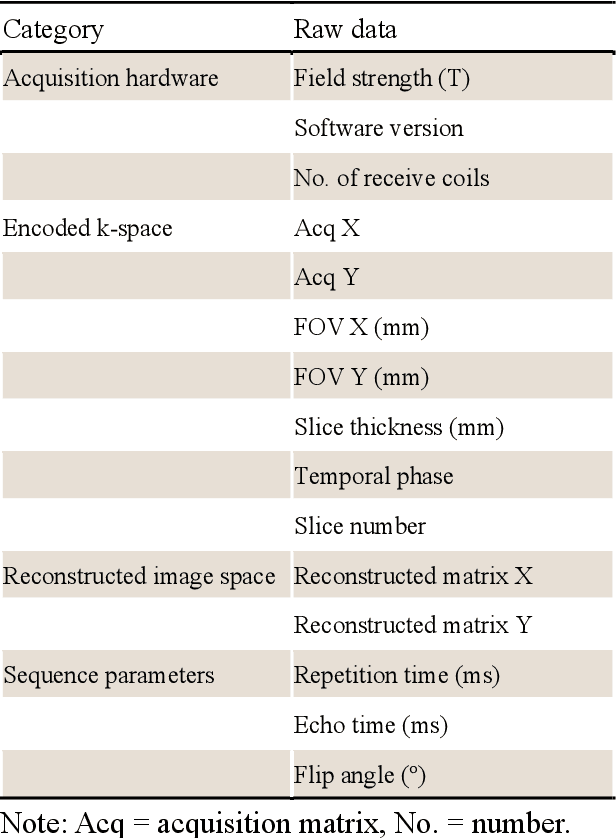
Abstract:Cardiac magnetic resonance imaging (MRI) has emerged as a clinically gold-standard technique for diagnosing cardiac diseases, thanks to its ability to provide diverse information with multiple modalities and anatomical views. Accelerated cardiac MRI is highly expected to achieve time-efficient and patient-friendly imaging, and then advanced image reconstruction approaches are required to recover high-quality, clinically interpretable images from undersampled measurements. However, the lack of publicly available cardiac MRI k-space dataset in terms of both quantity and diversity has severely hindered substantial technological progress, particularly for data-driven artificial intelligence. Here, we provide a standardized, diverse, and high-quality CMRxRecon2024 dataset to facilitate the technical development, fair evaluation, and clinical transfer of cardiac MRI reconstruction approaches, towards promoting the universal frameworks that enable fast and robust reconstructions across different cardiac MRI protocols in clinical practice. To the best of our knowledge, the CMRxRecon2024 dataset is the largest and most diverse publicly available cardiac k-space dataset. It is acquired from 330 healthy volunteers, covering commonly used modalities, anatomical views, and acquisition trajectories in clinical cardiac MRI workflows. Besides, an open platform with tutorials, benchmarks, and data processing tools is provided to facilitate data usage, advanced method development, and fair performance evaluation.
The state-of-the-art in Cardiac MRI Reconstruction: Results of the CMRxRecon Challenge in MICCAI 2023
Apr 01, 2024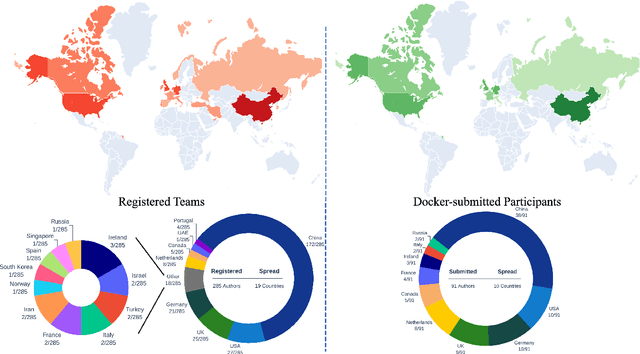
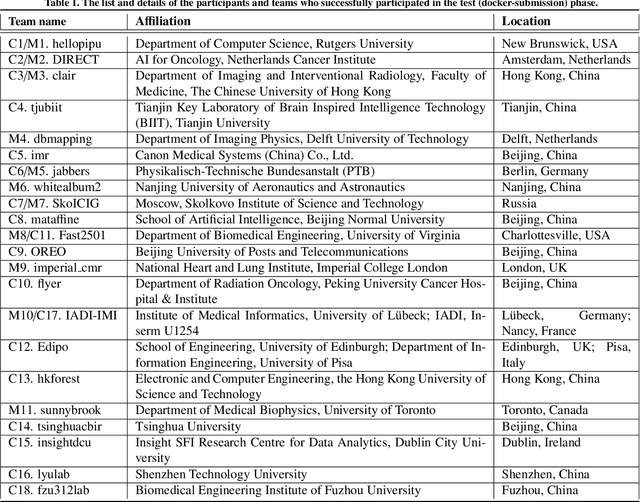
Abstract:Cardiac MRI, crucial for evaluating heart structure and function, faces limitations like slow imaging and motion artifacts. Undersampling reconstruction, especially data-driven algorithms, has emerged as a promising solution to accelerate scans and enhance imaging performance using highly under-sampled data. Nevertheless, the scarcity of publicly available cardiac k-space datasets and evaluation platform hinder the development of data-driven reconstruction algorithms. To address this issue, we organized the Cardiac MRI Reconstruction Challenge (CMRxRecon) in 2023, in collaboration with the 26th International Conference on MICCAI. CMRxRecon presented an extensive k-space dataset comprising cine and mapping raw data, accompanied by detailed annotations of cardiac anatomical structures. With overwhelming participation, the challenge attracted more than 285 teams and over 600 participants. Among them, 22 teams successfully submitted Docker containers for the testing phase, with 7 teams submitted for both cine and mapping tasks. All teams use deep learning based approaches, indicating that deep learning has predominately become a promising solution for the problem. The first-place winner of both tasks utilizes the E2E-VarNet architecture as backbones. In contrast, U-Net is still the most popular backbone for both multi-coil and single-coil reconstructions. This paper provides a comprehensive overview of the challenge design, presents a summary of the submitted results, reviews the employed methods, and offers an in-depth discussion that aims to inspire future advancements in cardiac MRI reconstruction models. The summary emphasizes the effective strategies observed in Cardiac MRI reconstruction, including backbone architecture, loss function, pre-processing techniques, physical modeling, and model complexity, thereby providing valuable insights for further developments in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge