Wu Yuan
Interpretable Droplet Digital PCR Assay for Trustworthy Molecular Diagnostics
Jan 16, 2025



Abstract:Accurate molecular quantification is essential for advancing research and diagnostics in fields such as infectious diseases, cancer biology, and genetic disorders. Droplet digital PCR (ddPCR) has emerged as a gold standard for achieving absolute quantification. While computational ddPCR technologies have advanced significantly, achieving automatic interpretation and consistent adaptability across diverse operational environments remains a challenge. To address these limitations, we introduce the intelligent interpretable droplet digital PCR (I2ddPCR) assay, a comprehensive framework integrating front-end predictive models (for droplet segmentation and classification) with GPT-4o multimodal large language model (MLLM, for context-aware explanations and recommendations) to automate and enhance ddPCR image analysis. This approach surpasses the state-of-the-art models, affording 99.05% accuracy in processing complex ddPCR images containing over 300 droplets per image with varying signal-to-noise ratios (SNRs). By combining specialized neural networks and large language models, the I2ddPCR assay offers a robust and adaptable solution for absolute molecular quantification, achieving a sensitivity capable of detecting low-abundance targets as low as 90.32 copies/{\mu}L. Furthermore, it improves model's transparency through detailed explanation and troubleshooting guidance, empowering users to make informed decisions. This innovative framework has the potential to benefit molecular diagnostics, disease research, and clinical applications, especially in resource-constrained settings.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Artificial Intelligence for Biomedical Video Generation
Nov 12, 2024



Abstract:As a prominent subfield of Artificial Intelligence Generated Content (AIGC), video generation has achieved notable advancements in recent years. The introduction of Sora-alike models represents a pivotal breakthrough in video generation technologies, significantly enhancing the quality of synthesized videos. Particularly in the realm of biomedicine, video generation technology has shown immense potential such as medical concept explanation, disease simulation, and biomedical data augmentation. In this article, we thoroughly examine the latest developments in video generation models and explore their applications, challenges, and future opportunities in the biomedical sector. We have conducted an extensive review and compiled a comprehensive list of datasets from various sources to facilitate the development and evaluation of video generative models in biomedicine. Given the rapid progress in this field, we have also created a github repository to regularly update the advances of biomedical video generation at: https://github.com/Lee728243228/Biomedical-Video-Generation
Fine-grained Classification of Port Wine Stains Using Optical Coherence Tomography Angiography
Aug 29, 2024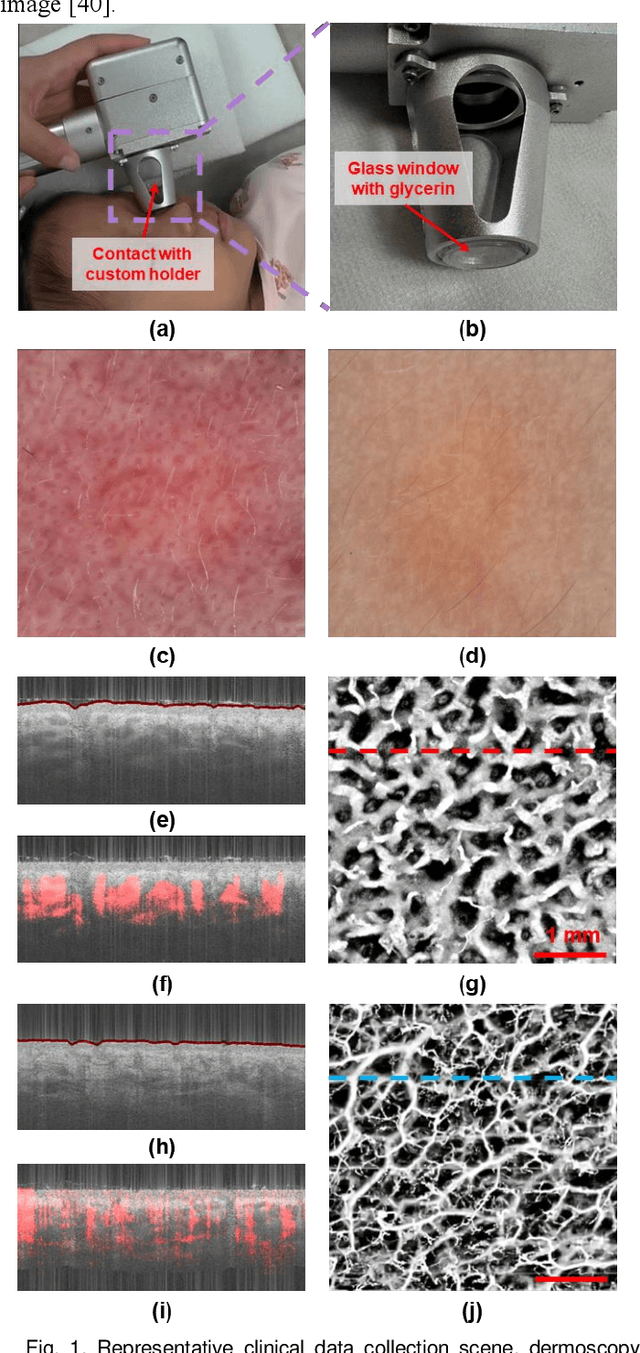
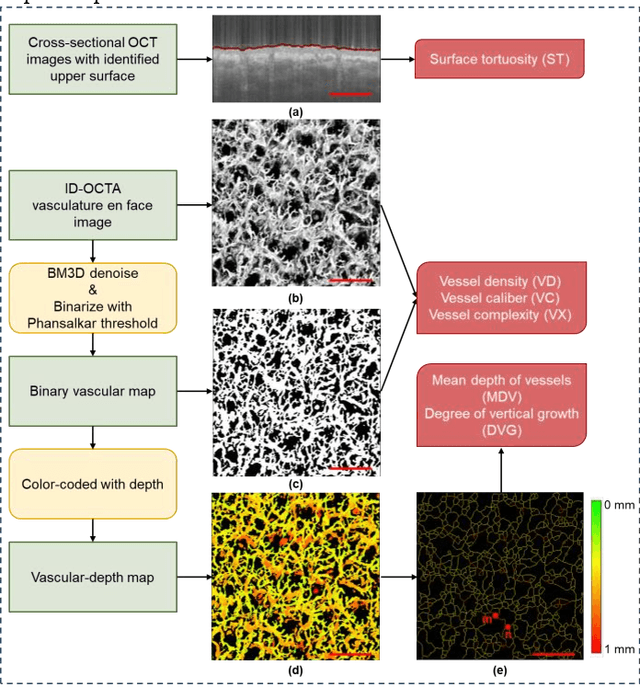

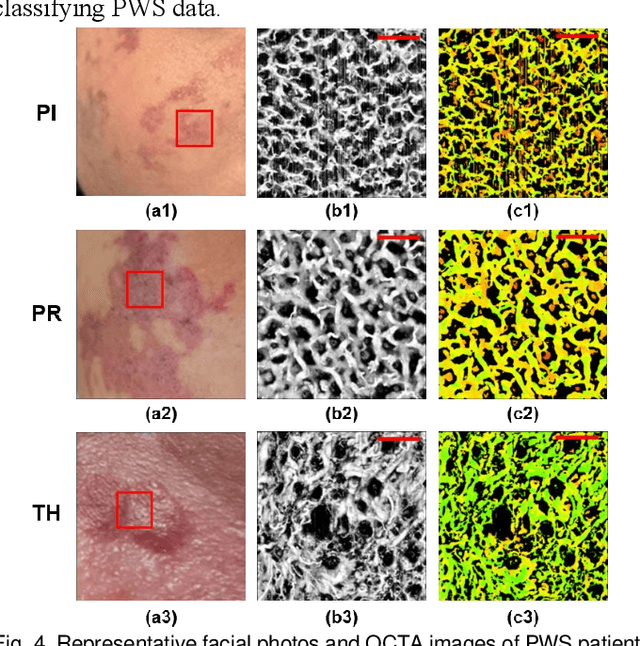
Abstract:Accurate classification of port wine stains (PWS, vascular malformations present at birth), is critical for subsequent treatment planning. However, the current method of classifying PWS based on the external skin appearance rarely reflects the underlying angiopathological heterogeneity of PWS lesions, resulting in inconsistent outcomes with the common vascular-targeted photodynamic therapy (V-PDT) treatments. Conversely, optical coherence tomography angiography (OCTA) is an ideal tool for visualizing the vascular malformations of PWS. Previous studies have shown no significant correlation between OCTA quantitative metrics and the PWS subtypes determined by the current classification approach. This study proposes a new classification approach for PWS using both OCT and OCTA. By examining the hypodermic histopathology and vascular structure of PWS, we have devised a fine-grained classification method that subdivides PWS into five distinct types. To assess the angiopathological differences of various PWS subtypes, we have analyzed six metrics related to vascular morphology and depth information of PWS lesions. The five PWS types present significant differences across all metrics compared to the conventional subtypes. Our findings suggest that an angiopathology-based classification accurately reflects the heterogeneity in PWS lesions. This research marks the first attempt to classify PWS based on angiopathology, potentially guiding more effective subtyping and treatment strategies for PWS.
MEDCO: Medical Education Copilots Based on A Multi-Agent Framework
Aug 22, 2024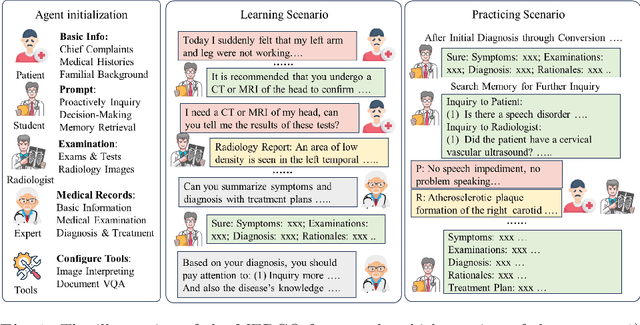
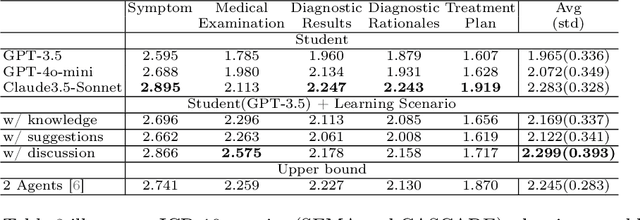

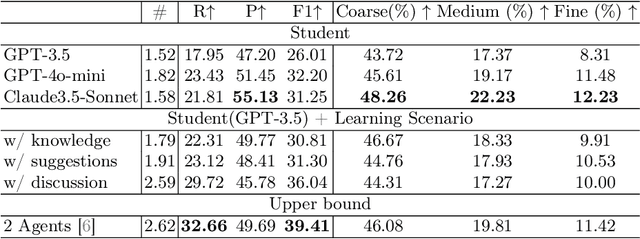
Abstract:Large language models (LLMs) have had a significant impact on diverse research domains, including medicine and healthcare. However, the potential of LLMs as copilots in medical education remains underexplored. Current AI-assisted educational tools are limited by their solitary learning approach and inability to simulate the multi-disciplinary and interactive nature of actual medical training. To address these limitations, we propose MEDCO (Medical EDucation COpilots), a novel multi-agent-based copilot system specially developed to emulate real-world medical training environments. MEDCO incorporates three primary agents: an agentic patient, an expert doctor, and a radiologist, facilitating a multi-modal and interactive learning environment. Our framework emphasizes the learning of proficient question-asking skills, multi-disciplinary collaboration, and peer discussions between students. Our experiments show that simulated virtual students who underwent training with MEDCO not only achieved substantial performance enhancements comparable to those of advanced models, but also demonstrated human-like learning behaviors and improvements, coupled with an increase in the number of learning samples. This work contributes to medical education by introducing a copilot that implements an interactive and collaborative learning approach. It also provides valuable insights into the effectiveness of AI-integrated training paradigms.
Artificial Intelligence Enhanced Digital Nucleic Acid Amplification Testing for Precision Medicine and Molecular Diagnostics
Jul 30, 2024



Abstract:The precise quantification of nucleic acids is pivotal in molecular biology, underscored by the rising prominence of nucleic acid amplification tests (NAAT) in diagnosing infectious diseases and conducting genomic studies. This review examines recent advancements in digital Polymerase Chain Reaction (dPCR) and digital Loop-mediated Isothermal Amplification (dLAMP), which surpass the limitations of traditional NAAT by offering absolute quantification and enhanced sensitivity. In this review, we summarize the compelling advancements of dNNAT in addressing pressing public health issues, especially during the COVID-19 pandemic. Further, we explore the transformative role of artificial intelligence (AI) in enhancing dNAAT image analysis, which not only improves efficiency and accuracy but also addresses traditional constraints related to cost, complexity, and data interpretation. In encompassing the state-of-the-art (SOTA) development and potential of both software and hardware, the all-encompassing Point-of-Care Testing (POCT) systems cast new light on benefits including higher throughput, label-free detection, and expanded multiplex analyses. While acknowledging the enhancement of AI-enhanced dNAAT technology, this review aims to both fill critical gaps in the existing technologies through comparative assessments and offer a balanced perspective on the current trajectory, including attendant challenges and future directions. Leveraging AI, next-generation dPCR and dLAMP technologies promises integration into clinical practice, improving personalized medicine, real-time epidemic surveillance, and global diagnostic accessibility.
VisionCLIP: An Med-AIGC based Ethical Language-Image Foundation Model for Generalizable Retina Image Analysis
Mar 16, 2024Abstract:Generalist foundation model has ushered in newfound capabilities in medical domain. However, the contradiction between the growing demand for high-quality annotated data with patient privacy continues to intensify. The utilization of medical artificial intelligence generated content (Med-AIGC) as an inexhaustible resource repository arises as a potential solution to address the aforementioned challenge. Here we harness 1 million open-source synthetic fundus images paired with natural language descriptions, to curate an ethical language-image foundation model for retina image analysis named VisionCLIP. VisionCLIP achieves competitive performance on three external datasets compared with the existing method pre-trained on real-world data in a zero-shot fashion. The employment of artificially synthetic images alongside corresponding textual data for training enables the medical foundation model to successfully assimilate knowledge of disease symptomatology, thereby circumventing potential breaches of patient confidentiality.
A Survey of Reasoning with Foundation Models
Dec 26, 2023

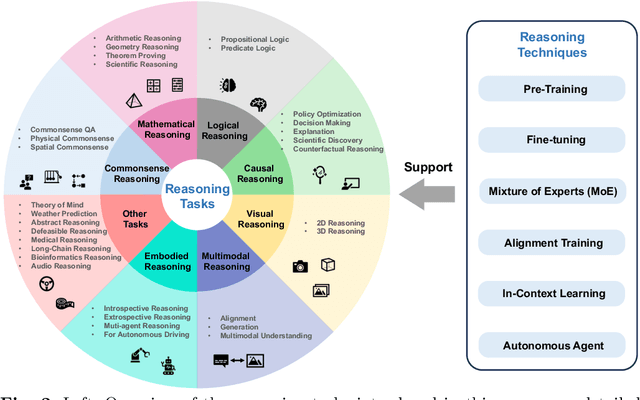
Abstract:Reasoning, a crucial ability for complex problem-solving, plays a pivotal role in various real-world settings such as negotiation, medical diagnosis, and criminal investigation. It serves as a fundamental methodology in the field of Artificial General Intelligence (AGI). With the ongoing development of foundation models, there is a growing interest in exploring their abilities in reasoning tasks. In this paper, we introduce seminal foundation models proposed or adaptable for reasoning, highlighting the latest advancements in various reasoning tasks, methods, and benchmarks. We then delve into the potential future directions behind the emergence of reasoning abilities within foundation models. We also discuss the relevance of multimodal learning, autonomous agents, and super alignment in the context of reasoning. By discussing these future research directions, we hope to inspire researchers in their exploration of this field, stimulate further advancements in reasoning with foundation models, and contribute to the development of AGI.
Dietary Assessment with Multimodal ChatGPT: A Systematic Analysis
Dec 14, 2023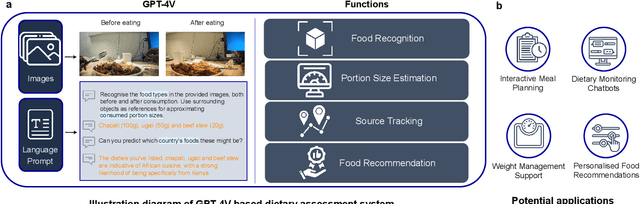

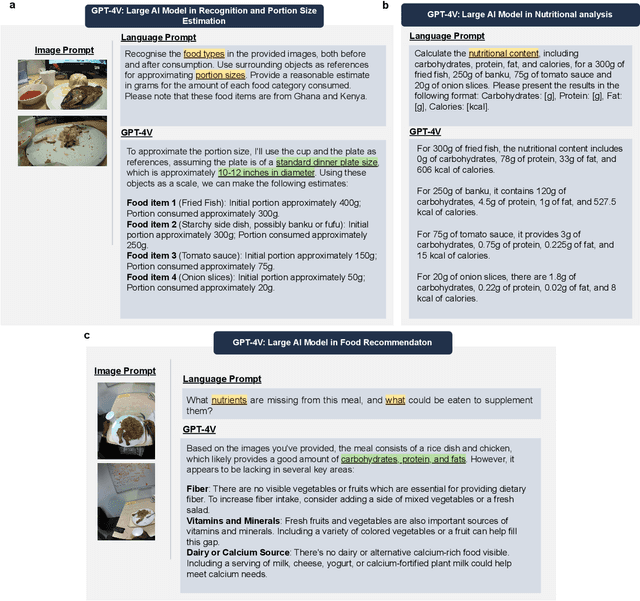

Abstract:Conventional approaches to dietary assessment are primarily grounded in self-reporting methods or structured interviews conducted under the supervision of dietitians. These methods, however, are often subjective, potentially inaccurate, and time-intensive. Although artificial intelligence (AI)-based solutions have been devised to automate the dietary assessment process, these prior AI methodologies encounter challenges in their ability to generalize across a diverse range of food types, dietary behaviors, and cultural contexts. This results in AI applications in the dietary field that possess a narrow specialization and limited accuracy. Recently, the emergence of multimodal foundation models such as GPT-4V powering the latest ChatGPT has exhibited transformative potential across a wide range of tasks (e.g., Scene understanding and image captioning) in numerous research domains. These models have demonstrated remarkable generalist intelligence and accuracy, capable of processing various data modalities. In this study, we explore the application of multimodal ChatGPT within the realm of dietary assessment. Our findings reveal that GPT-4V excels in food detection under challenging conditions with accuracy up to 87.5% without any fine-tuning or adaptation using food-specific datasets. By guiding the model with specific language prompts (e.g., African cuisine), it shifts from recognizing common staples like rice and bread to accurately identifying regional dishes like banku and ugali. Another GPT-4V's standout feature is its contextual awareness. GPT-4V can leverage surrounding objects as scale references to deduce the portion sizes of food items, further enhancing its accuracy in translating food weight into nutritional content. This alignment with the USDA National Nutrient Database underscores GPT-4V's potential to advance nutritional science and dietary assessment techniques.
Leveraging Multimodal Fusion for Enhanced Diagnosis of Multiple Retinal Diseases in Ultra-wide OCTA
Nov 17, 2023



Abstract:Ultra-wide optical coherence tomography angiography (UW-OCTA) is an emerging imaging technique that offers significant advantages over traditional OCTA by providing an exceptionally wide scanning range of up to 24 x 20 $mm^{2}$, covering both the anterior and posterior regions of the retina. However, the currently accessible UW-OCTA datasets suffer from limited comprehensive hierarchical information and corresponding disease annotations. To address this limitation, we have curated the pioneering M3OCTA dataset, which is the first multimodal (i.e., multilayer), multi-disease, and widest field-of-view UW-OCTA dataset. Furthermore, the effective utilization of multi-layer ultra-wide ocular vasculature information from UW-OCTA remains underdeveloped. To tackle this challenge, we propose the first cross-modal fusion framework that leverages multi-modal information for diagnosing multiple diseases. Through extensive experiments conducted on our openly available M3OCTA dataset, we demonstrate the effectiveness and superior performance of our method, both in fixed and varying modalities settings. The construction of the M3OCTA dataset, the first multimodal OCTA dataset encompassing multiple diseases, aims to advance research in the ophthalmic image analysis community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge