Yuanyuan Wei
MIRNet: Integrating Constrained Graph-Based Reasoning with Pre-training for Diagnostic Medical Imaging
Nov 13, 2025Abstract:Automated interpretation of medical images demands robust modeling of complex visual-semantic relationships while addressing annotation scarcity, label imbalance, and clinical plausibility constraints. We introduce MIRNet (Medical Image Reasoner Network), a novel framework that integrates self-supervised pre-training with constrained graph-based reasoning. Tongue image diagnosis is a particularly challenging domain that requires fine-grained visual and semantic understanding. Our approach leverages self-supervised masked autoencoder (MAE) to learn transferable visual representations from unlabeled data; employs graph attention networks (GAT) to model label correlations through expert-defined structured graphs; enforces clinical priors via constraint-aware optimization using KL divergence and regularization losses; and mitigates imbalance using asymmetric loss (ASL) and boosting ensembles. To address annotation scarcity, we also introduce TongueAtlas-4K, a comprehensive expert-curated benchmark comprising 4,000 images annotated with 22 diagnostic labels--representing the largest public dataset in tongue analysis. Validation shows our method achieves state-of-the-art performance. While optimized for tongue diagnosis, the framework readily generalizes to broader diagnostic medical imaging tasks.
An Integrated AI-Enabled System Using One Class Twin Cross Learning (OCT-X) for Early Gastric Cancer Detection
Mar 31, 2025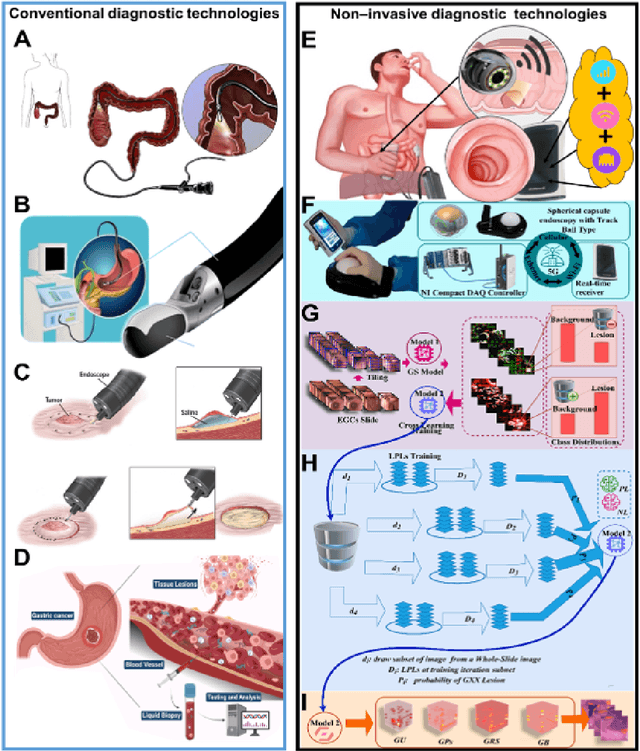
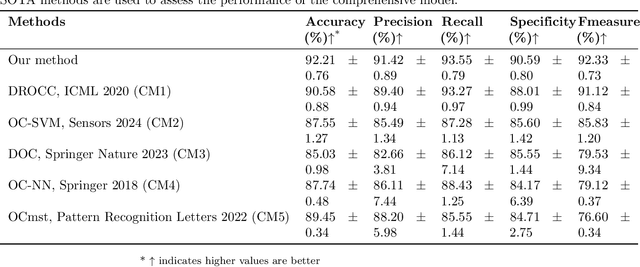
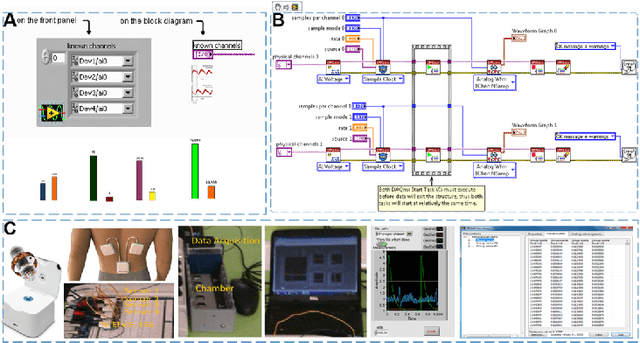
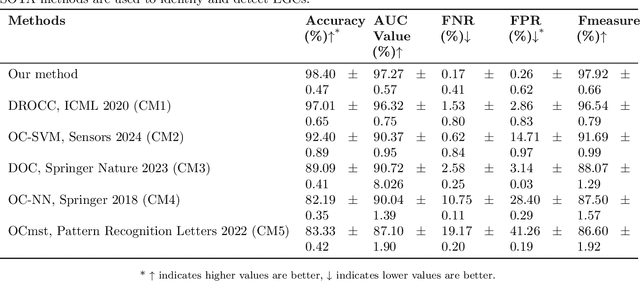
Abstract:Early detection of gastric cancer, a leading cause of cancer-related mortality worldwide, remains hampered by the limitations of current diagnostic technologies, leading to high rates of misdiagnosis and missed diagnoses. To address these challenges, we propose an integrated system that synergizes advanced hardware and software technologies to balance speed-accuracy. Our study introduces the One Class Twin Cross Learning (OCT-X) algorithm. Leveraging a novel fast double-threshold grid search strategy (FDT-GS) and a patch-based deep fully convolutional network, OCT-X maximizes diagnostic accuracy through real-time data processing and seamless lesion surveillance. The hardware component includes an all-in-one point-of-care testing (POCT) device with high-resolution imaging sensors, real-time data processing, and wireless connectivity, facilitated by the NI CompactDAQ and LabVIEW software. Our integrated system achieved an unprecedented diagnostic accuracy of 99.70%, significantly outperforming existing models by up to 4.47%, and demonstrated a 10% improvement in multirate adaptability. These findings underscore the potential of OCT-X as well as the integrated system in clinical diagnostics, offering a path toward more accurate, efficient, and less invasive early gastric cancer detection. Future research will explore broader applications, further advancing oncological diagnostics. Code is available at https://github.com/liu37972/Multirate-Location-on-OCT-X-Learning.git.
Interpretable Droplet Digital PCR Assay for Trustworthy Molecular Diagnostics
Jan 16, 2025



Abstract:Accurate molecular quantification is essential for advancing research and diagnostics in fields such as infectious diseases, cancer biology, and genetic disorders. Droplet digital PCR (ddPCR) has emerged as a gold standard for achieving absolute quantification. While computational ddPCR technologies have advanced significantly, achieving automatic interpretation and consistent adaptability across diverse operational environments remains a challenge. To address these limitations, we introduce the intelligent interpretable droplet digital PCR (I2ddPCR) assay, a comprehensive framework integrating front-end predictive models (for droplet segmentation and classification) with GPT-4o multimodal large language model (MLLM, for context-aware explanations and recommendations) to automate and enhance ddPCR image analysis. This approach surpasses the state-of-the-art models, affording 99.05% accuracy in processing complex ddPCR images containing over 300 droplets per image with varying signal-to-noise ratios (SNRs). By combining specialized neural networks and large language models, the I2ddPCR assay offers a robust and adaptable solution for absolute molecular quantification, achieving a sensitivity capable of detecting low-abundance targets as low as 90.32 copies/{\mu}L. Furthermore, it improves model's transparency through detailed explanation and troubleshooting guidance, empowering users to make informed decisions. This innovative framework has the potential to benefit molecular diagnostics, disease research, and clinical applications, especially in resource-constrained settings.
Enhancing Diagnostic Precision in Gastric Bleeding through Automated Lesion Segmentation: A Deep DuS-KFCM Approach
Nov 21, 2024


Abstract:Timely and precise classification and segmentation of gastric bleeding in endoscopic imagery are pivotal for the rapid diagnosis and intervention of gastric complications, which is critical in life-saving medical procedures. Traditional methods grapple with the challenge posed by the indistinguishable intensity values of bleeding tissues adjacent to other gastric structures. Our study seeks to revolutionize this domain by introducing a novel deep learning model, the Dual Spatial Kernelized Constrained Fuzzy C-Means (Deep DuS-KFCM) clustering algorithm. This Hybrid Neuro-Fuzzy system synergizes Neural Networks with Fuzzy Logic to offer a highly precise and efficient identification of bleeding regions. Implementing a two-fold coarse-to-fine strategy for segmentation, this model initially employs the Spatial Kernelized Fuzzy C-Means (SKFCM) algorithm enhanced with spatial intensity profiles and subsequently harnesses the state-of-the-art DeepLabv3+ with ResNet50 architecture to refine the segmentation output. Through extensive experiments across mainstream gastric bleeding and red spots datasets, our Deep DuS-KFCM model demonstrated unprecedented accuracy rates of 87.95%, coupled with a specificity of 96.33%, outperforming contemporary segmentation methods. The findings underscore the model's robustness against noise and its outstanding segmentation capabilities, particularly for identifying subtle bleeding symptoms, thereby presenting a significant leap forward in medical image processing.
Artificial Intelligence Enhanced Digital Nucleic Acid Amplification Testing for Precision Medicine and Molecular Diagnostics
Jul 30, 2024



Abstract:The precise quantification of nucleic acids is pivotal in molecular biology, underscored by the rising prominence of nucleic acid amplification tests (NAAT) in diagnosing infectious diseases and conducting genomic studies. This review examines recent advancements in digital Polymerase Chain Reaction (dPCR) and digital Loop-mediated Isothermal Amplification (dLAMP), which surpass the limitations of traditional NAAT by offering absolute quantification and enhanced sensitivity. In this review, we summarize the compelling advancements of dNNAT in addressing pressing public health issues, especially during the COVID-19 pandemic. Further, we explore the transformative role of artificial intelligence (AI) in enhancing dNAAT image analysis, which not only improves efficiency and accuracy but also addresses traditional constraints related to cost, complexity, and data interpretation. In encompassing the state-of-the-art (SOTA) development and potential of both software and hardware, the all-encompassing Point-of-Care Testing (POCT) systems cast new light on benefits including higher throughput, label-free detection, and expanded multiplex analyses. While acknowledging the enhancement of AI-enhanced dNAAT technology, this review aims to both fill critical gaps in the existing technologies through comparative assessments and offer a balanced perspective on the current trajectory, including attendant challenges and future directions. Leveraging AI, next-generation dPCR and dLAMP technologies promises integration into clinical practice, improving personalized medicine, real-time epidemic surveillance, and global diagnostic accessibility.
Auto-ICell: An Accessible and Cost-Effective Integrative Droplet Microfluidic System for Real-Time Single-Cell Morphological and Apoptotic Analysis
Nov 06, 2023Abstract:The Auto-ICell system, a novel, and cost-effective integrated droplet microfluidic system, is introduced for real-time analysis of single-cell morphology and apoptosis. This system integrates a 3D-printed microfluidic chip with image analysis algorithms, enabling the generation of uniform droplet reactors and immediate image analysis. The system employs a color-based image analysis algorithm in the bright field for droplet content analysis. Meanwhile, in the fluorescence field, cell apoptosis is quantitatively measured through a combination of deep-learning-enabled multiple fluorescent channel analysis and a live/dead cell stain kit. Breast cancer cells are encapsulated within uniform droplets, with diameters ranging from 70 {\mu}m to 240 {\mu}m, generated at a high throughput of 1,500 droplets per minute. Real-time image analysis results are displayed within 2 seconds on a custom graphical user interface (GUI). The system provides an automatic calculation of the distribution and ratio of encapsulated dyes in the bright field, and in the fluorescent field, cell blebbing and cell circularity are observed and quantified respectively. The Auto-ICell system is non-invasive and provides online detection, offering a robust, time-efficient, user-friendly, and cost-effective solution for single-cell analysis. It significantly enhances the detection throughput of droplet single-cell analysis by reducing setup costs and improving operational performance. This study highlights the potential of the Auto-ICell system in advancing biological research and personalized disease treatment, with promising applications in cell culture, biochemical microreactors, drug carriers, cell-based assays, synthetic biology, and point-of-care diagnostics.
Deep Learning Approach for Large-Scale, Real-Time Quantification of Green Fluorescent Protein-Labeled Biological Samples in Microreactors
Sep 04, 2023Abstract:Absolute quantification of biological samples entails determining expression levels in precise numerical copies, offering enhanced accuracy and superior performance for rare templates. However, existing methodologies suffer from significant limitations: flow cytometers are both costly and intricate, while fluorescence imaging relying on software tools or manual counting is time-consuming and prone to inaccuracies. In this study, we have devised a comprehensive deep-learning-enabled pipeline that enables the automated segmentation and classification of GFP (green fluorescent protein)-labeled microreactors, facilitating real-time absolute quantification. Our findings demonstrate the efficacy of this technique in accurately predicting the sizes and occupancy status of microreactors using standard laboratory fluorescence microscopes, thereby providing precise measurements of template concentrations. Notably, our approach exhibits an analysis speed of quantifying over 2,000 microreactors (across 10 images) within remarkably 2.5 seconds, and a dynamic range spanning from 56.52 to 1569.43 copies per micron-liter. Furthermore, our Deep-dGFP algorithm showcases remarkable generalization capabilities, as it can be directly applied to various GFP-labeling scenarios, including droplet-based, microwell-based, and agarose-based biological applications. To the best of our knowledge, this represents the first successful implementation of an all-in-one image analysis algorithm in droplet digital PCR (polymerase chain reaction), microwell digital PCR, droplet single-cell sequencing, agarose digital PCR, and bacterial quantification, without necessitating any transfer learning steps, modifications, or retraining procedures. We firmly believe that our Deep-dGFP technique will be readily embraced by biomedical laboratories and holds potential for further development in related clinical applications.
Lab-in-a-Tube: A portable imaging spectrophotometer for cost-effective, high-throughput, and label-free analysis of centrifugation processes
Aug 01, 2023



Abstract:Centrifuges serve as essential instruments in modern experimental sciences, facilitating a wide range of routine sample processing tasks that necessitate material sedimentation. However, the study for real time observation of the dynamical process during centrifugation has remained elusive. In this study, we developed an innovative Lab_in_a_Tube imaging spectrophotometer that incorporates capabilities of real time image analysis and programmable interruption. This portable LIAT device costs less than 30 US dollars. Based on our knowledge, it is the first Wi Fi camera built_in in common lab centrifuges with active closed_loop control. We tested our LIAT imaging spectrophotometer with solute solvent interaction investigation obtained from lab centrifuges with quantitative data plotting in a real time manner. Single re circulating flow was real time observed, forming the ring shaped pattern during centrifugation. To the best of our knowledge, this is the very first observation of similar phenomena. We developed theoretical simulations for the single particle in a rotating reference frame, which correlated well with experimental results. We also demonstrated the first demonstration to visualize the blood sedimentation process in clinical lab centrifuges. This remarkable cost effectiveness opens up exciting opportunities for centrifugation microbiology research and paves the way for the creation of a network of computational imaging spectrometers at an affordable price for large scale and continuous monitoring of centrifugal processes in general.
Classification and Explanation of Distributed Denial-of-Service (DDoS) Attack Detection using Machine Learning and Shapley Additive Explanation (SHAP) Methods
Jun 27, 2023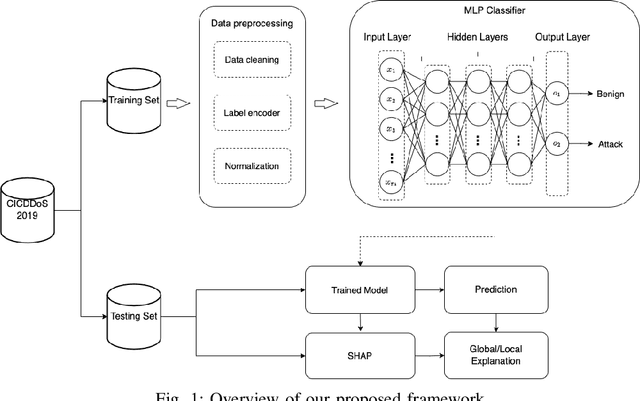
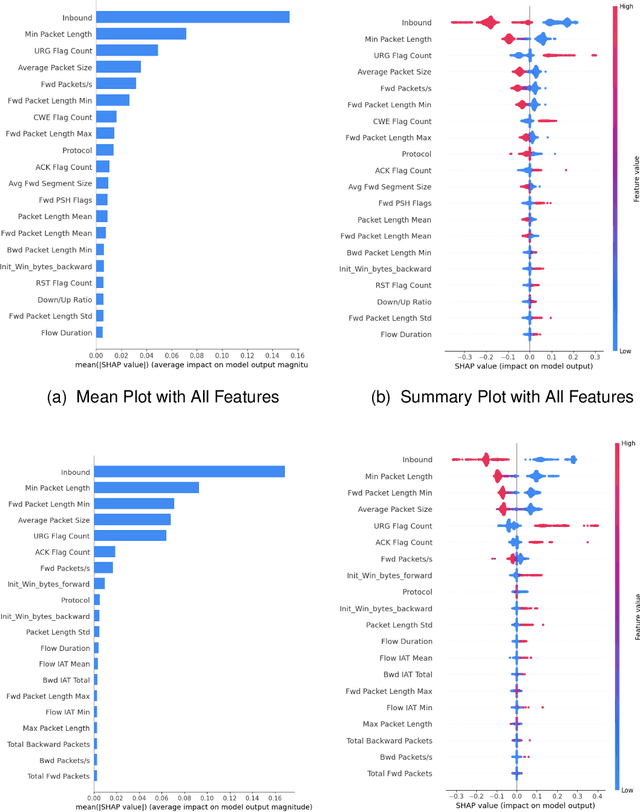
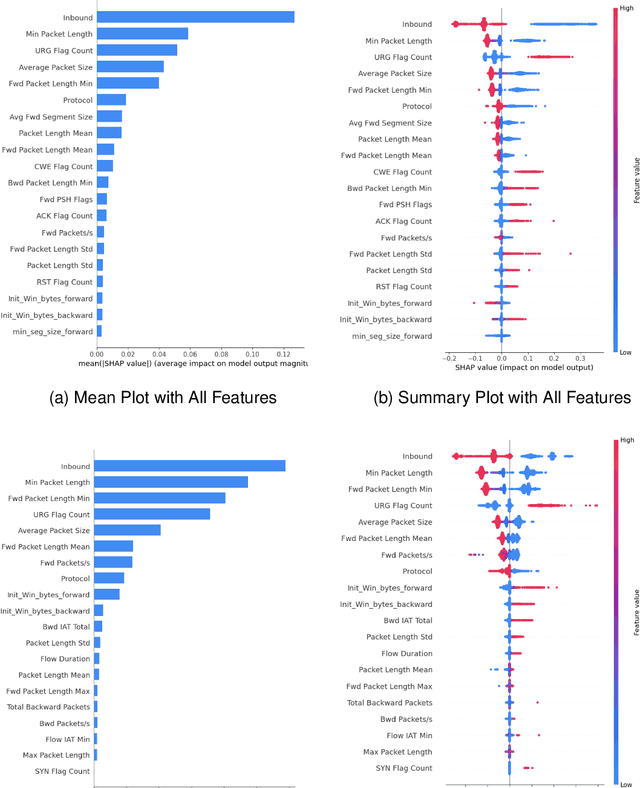
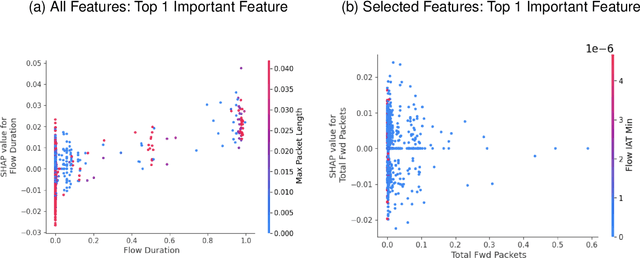
Abstract:DDoS attacks involve overwhelming a target system with a large number of requests or traffic from multiple sources, disrupting the normal traffic of a targeted server, service, or network. Distinguishing between legitimate traffic and malicious traffic is a challenging task. It is possible to classify legitimate traffic and malicious traffic and analysis the network traffic by using machine learning and deep learning techniques. However, an inter-model explanation implemented to classify a traffic flow whether is benign or malicious is an important investigation of the inner working theory of the model to increase the trustworthiness of the model. Explainable Artificial Intelligence (XAI) can explain the decision-making of the machine learning models that can be classified and identify DDoS traffic. In this context, we proposed a framework that can not only classify legitimate traffic and malicious traffic of DDoS attacks but also use SHAP to explain the decision-making of the classifier model. To address this concern, we first adopt feature selection techniques to select the top 20 important features based on feature importance techniques (e.g., XGB-based SHAP feature importance). Following that, the Multi-layer Perceptron Network (MLP) part of our proposed model uses the optimized features of the DDoS attack dataset as inputs to classify legitimate and malicious traffic. We perform extensive experiments with all features and selected features. The evaluation results show that the model performance with selected features achieves above 99\% accuracy. Finally, to provide interpretability, XAI can be adopted to explain the model performance between the prediction results and features based on global and local explanations by SHAP, which can better explain the results achieved by our proposed framework.
MProtoNet: A Case-Based Interpretable Model for Brain Tumor Classification with 3D Multi-parametric Magnetic Resonance Imaging
Apr 14, 2023



Abstract:Recent applications of deep convolutional neural networks in medical imaging raise concerns about their interpretability. While most explainable deep learning applications use post hoc methods (such as GradCAM) to generate feature attribution maps, there is a new type of case-based reasoning models, namely ProtoPNet and its variants, which identify prototypes during training and compare input image patches with those prototypes. We propose the first medical prototype network (MProtoNet) to extend ProtoPNet to brain tumor classification with 3D multi-parametric magnetic resonance imaging (mpMRI) data. To address different requirements between 2D natural images and 3D mpMRIs especially in terms of localizing attention regions, a new attention module with soft masking and online-CAM loss is introduced. Soft masking helps sharpen attention maps, while online-CAM loss directly utilizes image-level labels when training the attention module. MProtoNet achieves statistically significant improvements in interpretability metrics of both correctness and localization coherence (with a best activation precision of $0.713\pm0.058$) without human-annotated labels during training, when compared with GradCAM and several ProtoPNet variants. The source code is available at https://github.com/aywi/mprotonet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge