Sai Mu Dalike Abaxi
Deep Learning Approach for Large-Scale, Real-Time Quantification of Green Fluorescent Protein-Labeled Biological Samples in Microreactors
Sep 04, 2023Abstract:Absolute quantification of biological samples entails determining expression levels in precise numerical copies, offering enhanced accuracy and superior performance for rare templates. However, existing methodologies suffer from significant limitations: flow cytometers are both costly and intricate, while fluorescence imaging relying on software tools or manual counting is time-consuming and prone to inaccuracies. In this study, we have devised a comprehensive deep-learning-enabled pipeline that enables the automated segmentation and classification of GFP (green fluorescent protein)-labeled microreactors, facilitating real-time absolute quantification. Our findings demonstrate the efficacy of this technique in accurately predicting the sizes and occupancy status of microreactors using standard laboratory fluorescence microscopes, thereby providing precise measurements of template concentrations. Notably, our approach exhibits an analysis speed of quantifying over 2,000 microreactors (across 10 images) within remarkably 2.5 seconds, and a dynamic range spanning from 56.52 to 1569.43 copies per micron-liter. Furthermore, our Deep-dGFP algorithm showcases remarkable generalization capabilities, as it can be directly applied to various GFP-labeling scenarios, including droplet-based, microwell-based, and agarose-based biological applications. To the best of our knowledge, this represents the first successful implementation of an all-in-one image analysis algorithm in droplet digital PCR (polymerase chain reaction), microwell digital PCR, droplet single-cell sequencing, agarose digital PCR, and bacterial quantification, without necessitating any transfer learning steps, modifications, or retraining procedures. We firmly believe that our Deep-dGFP technique will be readily embraced by biomedical laboratories and holds potential for further development in related clinical applications.
Generalist Vision Foundation Models for Medical Imaging: A Case Study of Segment Anything Model on Zero-Shot Medical Segmentation
Apr 25, 2023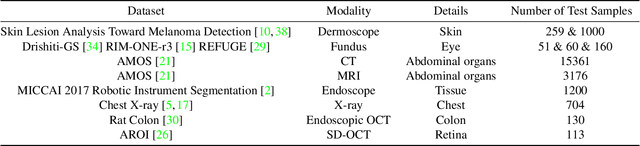
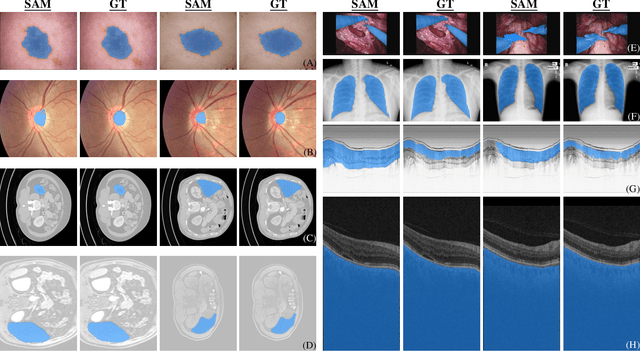
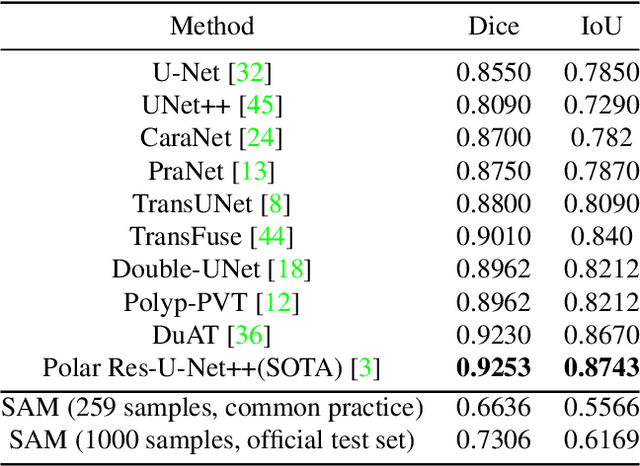
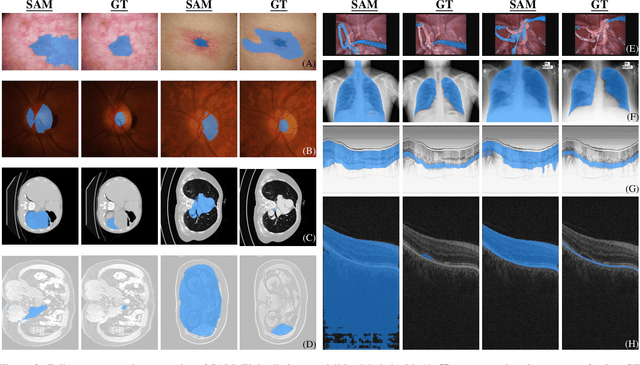
Abstract:We examine the recent Segment Anything Model (SAM) on medical images, and report both quantitative and qualitative zero-shot segmentation results on nine medical image segmentation benchmarks, covering various imaging modalities, such as optical coherence tomography (OCT), magnetic resonance imaging (MRI), and computed tomography (CT), as well as different applications including dermatology, ophthalmology, and radiology. Our experiments reveal that while SAM demonstrates stunning segmentation performance on images from the general domain, for those out-of-distribution images, e.g., medical images, its zero-shot segmentation performance is still limited. Furthermore, SAM demonstrated varying zero-shot segmentation performance across different unseen medical domains. For example, it had a 0.8704 mean Dice score on segmenting under-bruch's membrane layer of retinal OCT, whereas the segmentation accuracy drops to 0.0688 when segmenting retinal pigment epithelium. For certain structured targets, e.g., blood vessels, the zero-shot segmentation of SAM completely failed, whereas a simple fine-tuning of it with small amount of data could lead to remarkable improvements of the segmentation quality. Our study indicates the versatility of generalist vision foundation models on solving specific tasks in medical imaging, and their great potential to achieve desired performance through fine-turning and eventually tackle the challenges of accessing large diverse medical datasets and the complexity of medical domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge