Dehua Hu
A Two-Stage Adverse Weather Semantic Segmentation Method for WeatherProof Challenge CVPR 2024 Workshop UG2+
Jun 08, 2024Abstract:This technical report presents our team's solution for the WeatherProof Dataset Challenge: Semantic Segmentation in Adverse Weather at CVPR'24 UG2+. We propose a two-stage deep learning framework for this task. In the first stage, we preprocess the provided dataset by concatenating images into video sequences. Subsequently, we leverage a low-rank video deraining method to generate high-fidelity pseudo ground truths. These pseudo ground truths offer superior alignment compared to the original ground truths, facilitating model convergence during training. In the second stage, we employ the InternImage network to train for the semantic segmentation task using the generated pseudo ground truths. Notably, our meticulously designed framework demonstrates robustness to degraded data captured under adverse weather conditions. In the challenge, our solution achieved a competitive score of 0.43 on the Mean Intersection over Union (mIoU) metric, securing a respectable rank of 4th.
The Ninth NTIRE 2024 Efficient Super-Resolution Challenge Report
Apr 16, 2024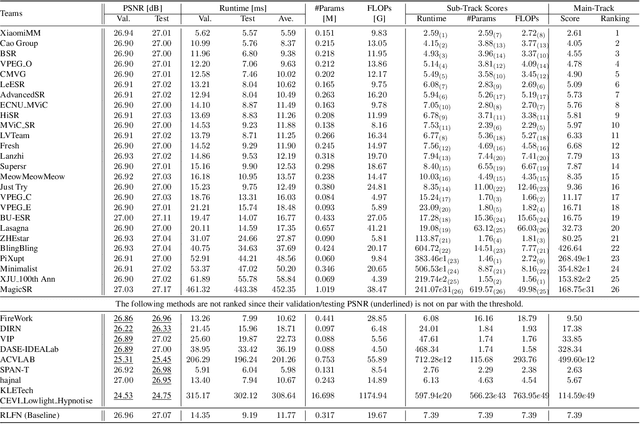
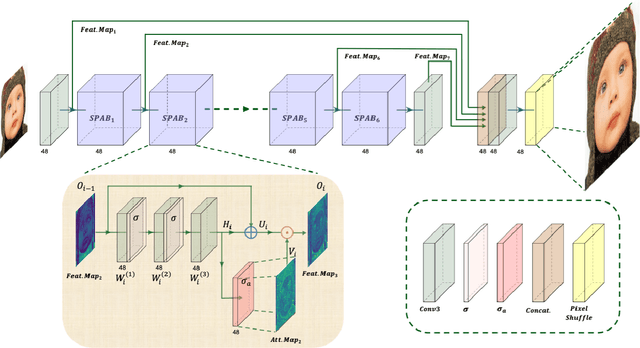
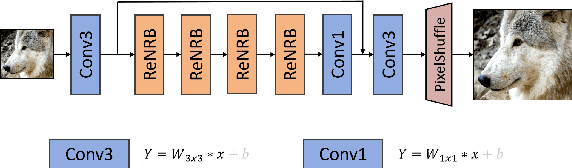

Abstract:This paper provides a comprehensive review of the NTIRE 2024 challenge, focusing on efficient single-image super-resolution (ESR) solutions and their outcomes. The task of this challenge is to super-resolve an input image with a magnification factor of x4 based on pairs of low and corresponding high-resolution images. The primary objective is to develop networks that optimize various aspects such as runtime, parameters, and FLOPs, while still maintaining a peak signal-to-noise ratio (PSNR) of approximately 26.90 dB on the DIV2K_LSDIR_valid dataset and 26.99 dB on the DIV2K_LSDIR_test dataset. In addition, this challenge has 4 tracks including the main track (overall performance), sub-track 1 (runtime), sub-track 2 (FLOPs), and sub-track 3 (parameters). In the main track, all three metrics (ie runtime, FLOPs, and parameter count) were considered. The ranking of the main track is calculated based on a weighted sum-up of the scores of all other sub-tracks. In sub-track 1, the practical runtime performance of the submissions was evaluated, and the corresponding score was used to determine the ranking. In sub-track 2, the number of FLOPs was considered. The score calculated based on the corresponding FLOPs was used to determine the ranking. In sub-track 3, the number of parameters was considered. The score calculated based on the corresponding parameters was used to determine the ranking. RLFN is set as the baseline for efficiency measurement. The challenge had 262 registered participants, and 34 teams made valid submissions. They gauge the state-of-the-art in efficient single-image super-resolution. To facilitate the reproducibility of the challenge and enable other researchers to build upon these findings, the code and the pre-trained model of validated solutions are made publicly available at https://github.com/Amazingren/NTIRE2024_ESR/.
Deep Learning Approach for Large-Scale, Real-Time Quantification of Green Fluorescent Protein-Labeled Biological Samples in Microreactors
Sep 04, 2023Abstract:Absolute quantification of biological samples entails determining expression levels in precise numerical copies, offering enhanced accuracy and superior performance for rare templates. However, existing methodologies suffer from significant limitations: flow cytometers are both costly and intricate, while fluorescence imaging relying on software tools or manual counting is time-consuming and prone to inaccuracies. In this study, we have devised a comprehensive deep-learning-enabled pipeline that enables the automated segmentation and classification of GFP (green fluorescent protein)-labeled microreactors, facilitating real-time absolute quantification. Our findings demonstrate the efficacy of this technique in accurately predicting the sizes and occupancy status of microreactors using standard laboratory fluorescence microscopes, thereby providing precise measurements of template concentrations. Notably, our approach exhibits an analysis speed of quantifying over 2,000 microreactors (across 10 images) within remarkably 2.5 seconds, and a dynamic range spanning from 56.52 to 1569.43 copies per micron-liter. Furthermore, our Deep-dGFP algorithm showcases remarkable generalization capabilities, as it can be directly applied to various GFP-labeling scenarios, including droplet-based, microwell-based, and agarose-based biological applications. To the best of our knowledge, this represents the first successful implementation of an all-in-one image analysis algorithm in droplet digital PCR (polymerase chain reaction), microwell digital PCR, droplet single-cell sequencing, agarose digital PCR, and bacterial quantification, without necessitating any transfer learning steps, modifications, or retraining procedures. We firmly believe that our Deep-dGFP technique will be readily embraced by biomedical laboratories and holds potential for further development in related clinical applications.
Lab-in-a-Tube: A portable imaging spectrophotometer for cost-effective, high-throughput, and label-free analysis of centrifugation processes
Aug 01, 2023



Abstract:Centrifuges serve as essential instruments in modern experimental sciences, facilitating a wide range of routine sample processing tasks that necessitate material sedimentation. However, the study for real time observation of the dynamical process during centrifugation has remained elusive. In this study, we developed an innovative Lab_in_a_Tube imaging spectrophotometer that incorporates capabilities of real time image analysis and programmable interruption. This portable LIAT device costs less than 30 US dollars. Based on our knowledge, it is the first Wi Fi camera built_in in common lab centrifuges with active closed_loop control. We tested our LIAT imaging spectrophotometer with solute solvent interaction investigation obtained from lab centrifuges with quantitative data plotting in a real time manner. Single re circulating flow was real time observed, forming the ring shaped pattern during centrifugation. To the best of our knowledge, this is the very first observation of similar phenomena. We developed theoretical simulations for the single particle in a rotating reference frame, which correlated well with experimental results. We also demonstrated the first demonstration to visualize the blood sedimentation process in clinical lab centrifuges. This remarkable cost effectiveness opens up exciting opportunities for centrifugation microbiology research and paves the way for the creation of a network of computational imaging spectrometers at an affordable price for large scale and continuous monitoring of centrifugal processes in general.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge