Lauren J. O'Donnell
Cross-Population White Matter Atlas Creation for Concurrent Mapping of Brain Connections in Neonates and Adults with Diffusion MRI Tractography
Dec 23, 2025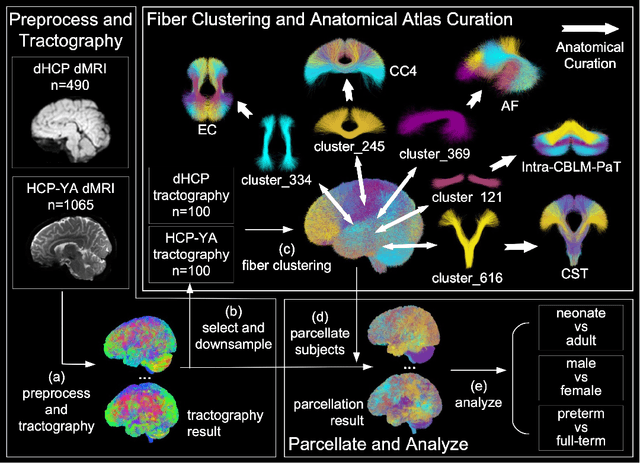
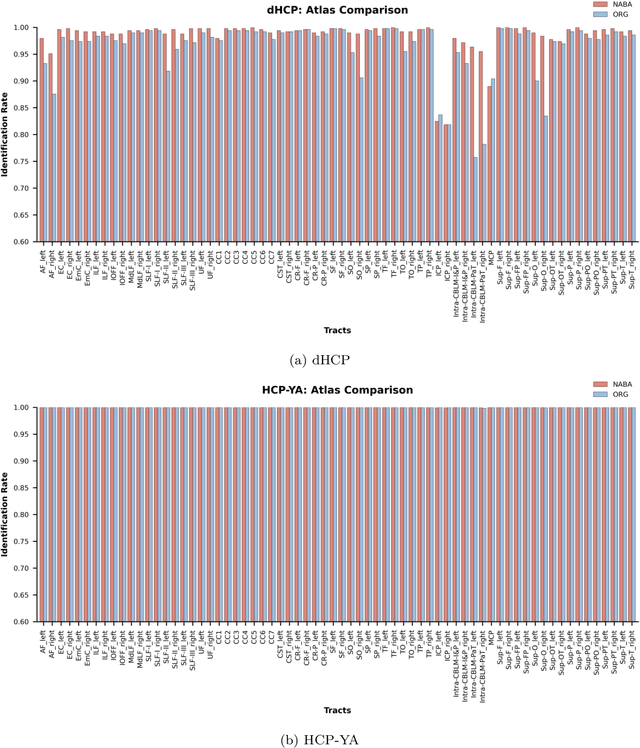
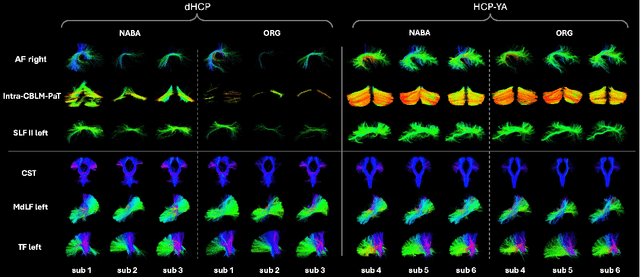
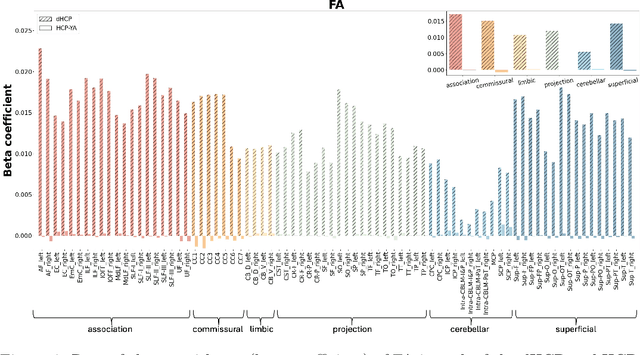
Abstract:Comparing white matter (WM) connections between adults and neonates using diffusion MRI (dMRI) can advance our understanding of typical brain development and potential biomarkers for neurological disorders. However, existing WM atlases are population-specific (adult or neonatal) and reside in separate spaces, preventing direct cross-population comparisons. A unified WM atlas spanning both neonates and adults is still lacking. In this study, we propose a neonatal/adult brain atlas (NABA), a WM tractography atlas built from dMRI data of both neonates and adults. NABA is constructed using a robust, data-driven fiber clustering pipeline, enabling group-wise WM atlasing across populations despite substantial anatomical variability. The atlas provides a standardized template for WM parcellation, allowing direct comparison of WM tracts between neonates and adults. Using NABA, we conduct four analyses: (1) evaluating the feasibility of joint WM mapping across populations, (2) characterizing WM development across neonatal ages relative to adults, (3) assessing sex-related differences in neonatal WM development, and (4) examining the effects of preterm birth. Our results show that NABA robustly identifies WM tracts in both populations. We observe rapid fractional anisotropy (FA) development in long-range association tracts, including the arcuate fasciculus and superior longitudinal fasciculus II, whereas intra-cerebellar tracts develop more slowly. Neonatal females exhibit faster overall FA development than males. Although preterm neonates show lower overall FA development rates, they demonstrate relatively higher FA growth in specific tracts, including the corticospinal tract, corona radiata-pontine pathway, and intracerebellar tracts. These findings demonstrate that NABA is a useful tool for investigating WM development across neonates and adults.
DDTracking: A Deep Generative Framework for Diffusion MRI Tractography with Streamline Local-Global Spatiotemporal Modeling
Aug 06, 2025Abstract:This paper presents DDTracking, a novel deep generative framework for diffusion MRI tractography that formulates streamline propagation as a conditional denoising diffusion process. In DDTracking, we introduce a dual-pathway encoding network that jointly models local spatial encoding (capturing fine-scale structural details at each streamline point) and global temporal dependencies (ensuring long-range consistency across the entire streamline). Furthermore, we design a conditional diffusion model module, which leverages the learned local and global embeddings to predict streamline propagation orientations for tractography in an end-to-end trainable manner. We conduct a comprehensive evaluation across diverse, independently acquired dMRI datasets, including both synthetic and clinical data. Experiments on two well-established benchmarks with ground truth (ISMRM Challenge and TractoInferno) demonstrate that DDTracking largely outperforms current state-of-the-art tractography methods. Furthermore, our results highlight DDTracking's strong generalizability across heterogeneous datasets, spanning varying health conditions, age groups, imaging protocols, and scanner types. Collectively, DDTracking offers anatomically plausible and robust tractography, presenting a scalable, adaptable, and end-to-end learnable solution for broad dMRI applications. Code is available at: https://github.com/yishengpoxiao/DDtracking.git
DeepMultiConnectome: Deep Multi-Task Prediction of Structural Connectomes Directly from Diffusion MRI Tractography
May 27, 2025



Abstract:Diffusion MRI (dMRI) tractography enables in vivo mapping of brain structural connections, but traditional connectome generation is time-consuming and requires gray matter parcellation, posing challenges for large-scale studies. We introduce DeepMultiConnectome, a deep-learning model that predicts structural connectomes directly from tractography, bypassing the need for gray matter parcellation while supporting multiple parcellation schemes. Using a point-cloud-based neural network with multi-task learning, the model classifies streamlines according to their connected regions across two parcellation schemes, sharing a learned representation. We train and validate DeepMultiConnectome on tractography from the Human Connectome Project Young Adult dataset ($n = 1000$), labeled with an 84 and 164 region gray matter parcellation scheme. DeepMultiConnectome predicts multiple structural connectomes from a whole-brain tractogram containing 3 million streamlines in approximately 40 seconds. DeepMultiConnectome is evaluated by comparing predicted connectomes with traditional connectomes generated using the conventional method of labeling streamlines using a gray matter parcellation. The predicted connectomes are highly correlated with traditionally generated connectomes ($r = 0.992$ for an 84-region scheme; $r = 0.986$ for a 164-region scheme) and largely preserve network properties. A test-retest analysis of DeepMultiConnectome demonstrates reproducibility comparable to traditionally generated connectomes. The predicted connectomes perform similarly to traditionally generated connectomes in predicting age and cognitive function. Overall, DeepMultiConnectome provides a scalable, fast model for generating subject-specific connectomes across multiple parcellation schemes.
A Multimodal Deep Learning Approach for White Matter Shape Prediction in Diffusion MRI Tractography
Apr 25, 2025Abstract:Shape measures have emerged as promising descriptors of white matter tractography, offering complementary insights into anatomical variability and associations with cognitive and clinical phenotypes. However, conventional methods for computing shape measures are computationally expensive and time-consuming for large-scale datasets due to reliance on voxel-based representations. We propose Tract2Shape, a novel multimodal deep learning framework that leverages geometric (point cloud) and scalar (tabular) features to predict ten white matter tractography shape measures. To enhance model efficiency, we utilize a dimensionality reduction algorithm for the model to predict five primary shape components. The model is trained and evaluated on two independently acquired datasets, the HCP-YA dataset, and the PPMI dataset. We evaluate the performance of Tract2Shape by training and testing it on the HCP-YA dataset and comparing the results with state-of-the-art models. To further assess its robustness and generalization ability, we also test Tract2Shape on the unseen PPMI dataset. Tract2Shape outperforms SOTA deep learning models across all ten shape measures, achieving the highest average Pearson's r and the lowest nMSE on the HCP-YA dataset. The ablation study shows that both multimodal input and PCA contribute to performance gains. On the unseen testing PPMI dataset, Tract2Shape maintains a high Pearson's r and low nMSE, demonstrating strong generalizability in cross-dataset evaluation. Tract2Shape enables fast, accurate, and generalizable prediction of white matter shape measures from tractography data, supporting scalable analysis across datasets. This framework lays a promising foundation for future large-scale white matter shape analysis.
DeepNuParc: A Novel Deep Clustering Framework for Fine-scale Parcellation of Brain Nuclei Using Diffusion MRI Tractography
Mar 10, 2025



Abstract:Brain nuclei are clusters of anatomically distinct neurons that serve as important hubs for processing and relaying information in various neural circuits. Fine-scale parcellation of the brain nuclei is vital for a comprehensive understanding of its anatomico-functional correlations. Diffusion MRI tractography is an advanced imaging technique that can estimate the brain's white matter structural connectivity to potentially reveal the topography of the nuclei of interest for studying its subdivisions. In this work, we present a deep clustering pipeline, namely DeepNuParc, to perform automated, fine-scale parcellation of brain nuclei using diffusion MRI tractography. First, we incorporate a newly proposed deep learning approach to enable accurate segmentation of the nuclei of interest directly on the dMRI data. Next, we design a novel streamline clustering-based structural connectivity feature for a robust representation of voxels within the nuclei. Finally, we improve the popular joint dimensionality reduction and k-means clustering approach to enable nuclei parcellation at a finer scale. We demonstrate DeepNuParc on two important brain structures, i.e. the amygdala and the thalamus, that are known to have multiple anatomically and functionally distinct nuclei subdivisions. Experimental results show that DeepNuParc enables consistent parcellation of the nuclei into multiple parcels across multiple subjects and achieves good correspondence with the widely used coarse-scale atlases. Our codes are available at https://github.com/HarlandZZC/deep_nuclei_parcellation.
MultiCo3D: Multi-Label Voxel Contrast for One-Shot Incremental Segmentation of 3D Neuroimages
Mar 09, 2025Abstract:3D neuroimages provide a comprehensive view of brain structure and function, aiding in precise localization and functional connectivity analysis. Segmentation of white matter (WM) tracts using 3D neuroimages is vital for understanding the brain's structural connectivity in both healthy and diseased states. One-shot Class Incremental Semantic Segmentation (OCIS) refers to effectively segmenting new (novel) classes using only a single sample while retaining knowledge of old (base) classes without forgetting. Voxel-contrastive OCIS methods adjust the feature space to alleviate the feature overlap problem between the base and novel classes. However, since WM tract segmentation is a multi-label segmentation task, existing single-label voxel contrastive-based methods may cause inherent contradictions. To address this, we propose a new multi-label voxel contrast framework called MultiCo3D for one-shot class incremental tract segmentation. Our method utilizes uncertainty distillation to preserve base tract segmentation knowledge while adjusting the feature space with multi-label voxel contrast to alleviate feature overlap when learning novel tracts and dynamically weighting multi losses to balance overall loss. We compare our method against several state-of-the-art (SOTA) approaches. The experimental results show that our method significantly enhances one-shot class incremental tract segmentation accuracy across five different experimental setups on HCP and Preto datasets.
DDCSR: A Novel End-to-End Deep Learning Framework for Cortical Surface Reconstruction from Diffusion MRI
Mar 05, 2025Abstract:Diffusion MRI (dMRI) plays a crucial role in studying brain white matter connectivity. Cortical surface reconstruction (CSR), including the inner whiter matter (WM) and outer pial surfaces, is one of the key tasks in dMRI analyses such as fiber tractography and multimodal MRI analysis. Existing CSR methods rely on anatomical T1-weighted data and map them into the dMRI space through inter-modality registration. However, due to the low resolution and image distortions of dMRI data, inter-modality registration faces significant challenges. This work proposes a novel end-to-end learning framework, DDCSR, which for the first time enables CSR directly from dMRI data. DDCSR consists of two major components, including: (1) an implicit learning module to predict a voxel-wise intermediate surface representation, and (2) an explicit learning module to predict the 3D mesh surfaces. Compared to several baseline and advanced CSR methods, we show that the proposed DDCSR can largely increase both accuracy and efficiency. Furthermore, we demonstrate a high generalization ability of DDCSR to data from different sources, despite the differences in dMRI acquisitions and populations.
A Novel Streamline-based diffusion MRI Tractography Registration Method with Probabilistic Keypoint Detection
Mar 04, 2025


Abstract:Registration of diffusion MRI tractography is an essential step for analyzing group similarities and variations in the brain's white matter (WM). Streamline-based registration approaches can leverage the 3D geometric information of fiber pathways to enable spatial alignment after registration. Existing methods usually rely on the optimization of the spatial distances to identify the optimal transformation. However, such methods overlook point connectivity patterns within the streamline itself, limiting their ability to identify anatomical correspondences across tractography datasets. In this work, we propose a novel unsupervised approach using deep learning to perform streamline-based dMRI tractography registration. The overall idea is to identify corresponding keypoint pairs across subjects for spatial alignment of tractography datasets. We model tractography as point clouds to leverage the graph connectivity along streamlines. We propose a novel keypoint detection method for streamlines, framed as a probabilistic classification task to identify anatomically consistent correspondences across unstructured streamline sets. In the experiments, we compare several existing methods and show highly effective and efficient tractography registration performance.
TractCloud-FOV: Deep Learning-based Robust Tractography Parcellation in Diffusion MRI with Incomplete Field of View
Feb 28, 2025Abstract:Tractography parcellation classifies streamlines reconstructed from diffusion MRI into anatomically defined fiber tracts for clinical and research applications. However, clinical scans often have incomplete fields of view (FOV) where brain regions are partially imaged, leading to partial or truncated fiber tracts. To address this challenge, we introduce TractCloud-FOV, a deep learning framework that robustly parcellates tractography under conditions of incomplete FOV. We propose a novel training strategy, FOV-Cut Augmentation (FOV-CA), in which we synthetically cut tractograms to simulate a spectrum of real-world inferior FOV cutoff scenarios. This data augmentation approach enriches the training set with realistic truncated streamlines, enabling the model to achieve superior generalization. We evaluate the proposed TractCloud-FOV on both synthetically cut tractography and two real-life datasets with incomplete FOV. TractCloud-FOV significantly outperforms several state-of-the-art methods on all testing datasets in terms of streamline classification accuracy, generalization ability, tract anatomical depiction, and computational efficiency. Overall, TractCloud-FOV achieves efficient and consistent tractography parcellation in diffusion MRI with incomplete FOV.
Medical Image Registration Meets Vision Foundation Model: Prototype Learning and Contour Awareness
Feb 17, 2025Abstract:Medical image registration is a fundamental task in medical image analysis, aiming to establish spatial correspondences between paired images. However, existing unsupervised deformable registration methods rely solely on intensity-based similarity metrics, lacking explicit anatomical knowledge, which limits their accuracy and robustness. Vision foundation models, such as the Segment Anything Model (SAM), can generate high-quality segmentation masks that provide explicit anatomical structure knowledge, addressing the limitations of traditional methods that depend only on intensity similarity. Based on this, we propose a novel SAM-assisted registration framework incorporating prototype learning and contour awareness. The framework includes: (1) Explicit anatomical information injection, where SAM-generated segmentation masks are used as auxiliary inputs throughout training and testing to ensure the consistency of anatomical information; (2) Prototype learning, which leverages segmentation masks to extract prototype features and aligns prototypes to optimize semantic correspondences between images; and (3) Contour-aware loss, a contour-aware loss is designed that leverages the edges of segmentation masks to improve the model's performance in fine-grained deformation fields. Extensive experiments demonstrate that the proposed framework significantly outperforms existing methods across multiple datasets, particularly in challenging scenarios with complex anatomical structures and ambiguous boundaries. Our code is available at https://github.com/HaoXu0507/IPMI25-SAM-Assisted-Registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge