Jiapeng Zhang
LiNeXt: Revisiting LiDAR Completion with Efficient Non-Diffusion Architectures
Nov 13, 2025Abstract:3D LiDAR scene completion from point clouds is a fundamental component of perception systems in autonomous vehicles. Previous methods have predominantly employed diffusion models for high-fidelity reconstruction. However, their multi-step iterative sampling incurs significant computational overhead, limiting its real-time applicability. To address this, we propose LiNeXt-a lightweight, non-diffusion network optimized for rapid and accurate point cloud completion. Specifically, LiNeXt first applies the Noise-to-Coarse (N2C) Module to denoise the input noisy point cloud in a single pass, thereby obviating the multi-step iterative sampling of diffusion-based methods. The Refine Module then takes the coarse point cloud and its intermediate features from the N2C Module to perform more precise refinement, further enhancing structural completeness. Furthermore, we observe that LiDAR point clouds exhibit a distance-dependent spatial distribution, being densely sampled at proximal ranges and sparsely sampled at distal ranges. Accordingly, we propose the Distance-aware Selected Repeat strategy to generate a more uniformly distributed noisy point cloud. On the SemanticKITTI dataset, LiNeXt achieves a 199.8x speedup in inference, reduces Chamfer Distance by 50.7%, and uses only 6.1% of the parameters compared with LiDiff. These results demonstrate the superior efficiency and effectiveness of LiNeXt for real-time scene completion.
Enhancing Small-Scale Dataset Expansion with Triplet-Connection-based Sample Re-Weighting
Aug 11, 2025Abstract:The performance of computer vision models in certain real-world applications, such as medical diagnosis, is often limited by the scarcity of available images. Expanding datasets using pre-trained generative models is an effective solution. However, due to the uncontrollable generation process and the ambiguity of natural language, noisy images may be generated. Re-weighting is an effective way to address this issue by assigning low weights to such noisy images. We first theoretically analyze three types of supervision for the generated images. Based on the theoretical analysis, we develop TriReWeight, a triplet-connection-based sample re-weighting method to enhance generative data augmentation. Theoretically, TriReWeight can be integrated with any generative data augmentation methods and never downgrade their performance. Moreover, its generalization approaches the optimal in the order $O(\sqrt{d\ln (n)/n})$. Our experiments validate the correctness of the theoretical analysis and demonstrate that our method outperforms the existing SOTA methods by $7.9\%$ on average over six natural image datasets and by $3.4\%$ on average over three medical datasets. We also experimentally validate that our method can enhance the performance of different generative data augmentation methods.
A multi-core periphery perspective: Ranking via relative centrality
Jun 06, 2024Abstract:Community and core-periphery are two widely studied graph structures, with their coexistence observed in real-world graphs (Rombach, Porter, Fowler \& Mucha [SIAM J. App. Math. 2014, SIAM Review 2017]). However, the nature of this coexistence is not well understood and has been pointed out as an open problem (Yanchenko \& Sengupta [Statistics Surveys, 2023]). Especially, the impact of inferring the core-periphery structure of a graph on understanding its community structure is not well utilized. In this direction, we introduce a novel quantification for graphs with ground truth communities, where each community has a densely connected part (the core), and the rest is more sparse (the periphery), with inter-community edges more frequent between the peripheries. Built on this structure, we propose a new algorithmic concept that we call relative centrality to detect the cores. We observe that core-detection algorithms based on popular centrality measures such as PageRank and degree centrality can show some bias in their outcome by selecting very few vertices from some cores. We show that relative centrality solves this bias issue and provide theoretical and simulation support, as well as experiments on real-world graphs. Core detection is known to have important applications with respect to core-periphery structures. In our model, we show a new application: relative-centrality-based algorithms can select a subset of the vertices such that it contains sufficient vertices from all communities, and points in this subset are better separable into their respective communities. We apply the methods to 11 biological datasets, with our methods resulting in a more balanced selection of vertices from all communities such that clustering algorithms have better performance on this set.
FedMPQ: Secure and Communication-Efficient Federated Learning with Multi-codebook Product Quantization
Apr 21, 2024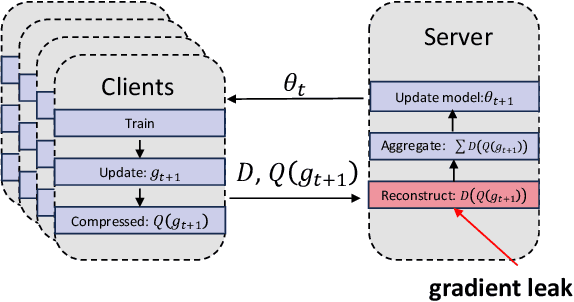

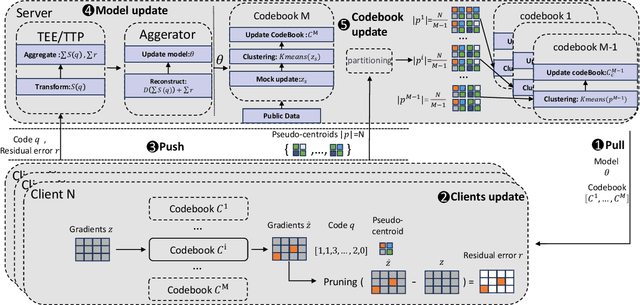
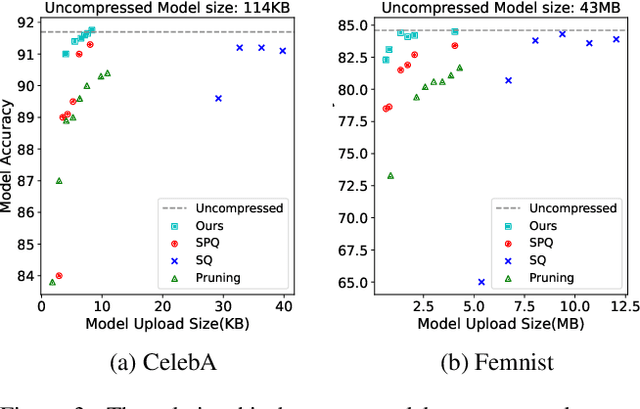
Abstract:In federated learning, particularly in cross-device scenarios, secure aggregation has recently gained popularity as it effectively defends against inference attacks by malicious aggregators. However, secure aggregation often requires additional communication overhead and can impede the convergence rate of the global model, which is particularly challenging in wireless network environments with extremely limited bandwidth. Therefore, achieving efficient communication compression under the premise of secure aggregation presents a highly challenging and valuable problem. In this work, we propose a novel uplink communication compression method for federated learning, named FedMPQ, which is based on multi shared codebook product quantization.Specifically, we utilize updates from the previous round to generate sufficiently robust codebooks. Secure aggregation is then achieved through trusted execution environments (TEE) or a trusted third party (TTP).In contrast to previous works, our approach exhibits greater robustness in scenarios where data is not independently and identically distributed (non-IID) and there is a lack of sufficient public data. The experiments conducted on the LEAF dataset demonstrate that our proposed method achieves 99% of the baseline's final accuracy, while reducing uplink communications by 90-95%
Deep Rib Fracture Instance Segmentation and Classification from CT on the RibFrac Challenge
Feb 14, 2024Abstract:Rib fractures are a common and potentially severe injury that can be challenging and labor-intensive to detect in CT scans. While there have been efforts to address this field, the lack of large-scale annotated datasets and evaluation benchmarks has hindered the development and validation of deep learning algorithms. To address this issue, the RibFrac Challenge was introduced, providing a benchmark dataset of over 5,000 rib fractures from 660 CT scans, with voxel-level instance mask annotations and diagnosis labels for four clinical categories (buckle, nondisplaced, displaced, or segmental). The challenge includes two tracks: a detection (instance segmentation) track evaluated by an FROC-style metric and a classification track evaluated by an F1-style metric. During the MICCAI 2020 challenge period, 243 results were evaluated, and seven teams were invited to participate in the challenge summary. The analysis revealed that several top rib fracture detection solutions achieved performance comparable or even better than human experts. Nevertheless, the current rib fracture classification solutions are hardly clinically applicable, which can be an interesting area in the future. As an active benchmark and research resource, the data and online evaluation of the RibFrac Challenge are available at the challenge website. As an independent contribution, we have also extended our previous internal baseline by incorporating recent advancements in large-scale pretrained networks and point-based rib segmentation techniques. The resulting FracNet+ demonstrates competitive performance in rib fracture detection, which lays a foundation for further research and development in AI-assisted rib fracture detection and diagnosis.
On the Power of SVD in the Stochastic Block Model
Sep 27, 2023Abstract:A popular heuristic method for improving clustering results is to apply dimensionality reduction before running clustering algorithms. It has been observed that spectral-based dimensionality reduction tools, such as PCA or SVD, improve the performance of clustering algorithms in many applications. This phenomenon indicates that spectral method not only serves as a dimensionality reduction tool, but also contributes to the clustering procedure in some sense. It is an interesting question to understand the behavior of spectral steps in clustering problems. As an initial step in this direction, this paper studies the power of vanilla-SVD algorithm in the stochastic block model (SBM). We show that, in the symmetric setting, vanilla-SVD algorithm recovers all clusters correctly. This result answers an open question posed by Van Vu (Combinatorics Probability and Computing, 2018) in the symmetric setting.
A Lightweight Domain Adversarial Neural Network Based on Knowledge Distillation for EEG-based Cross-subject Emotion Recognition
May 12, 2023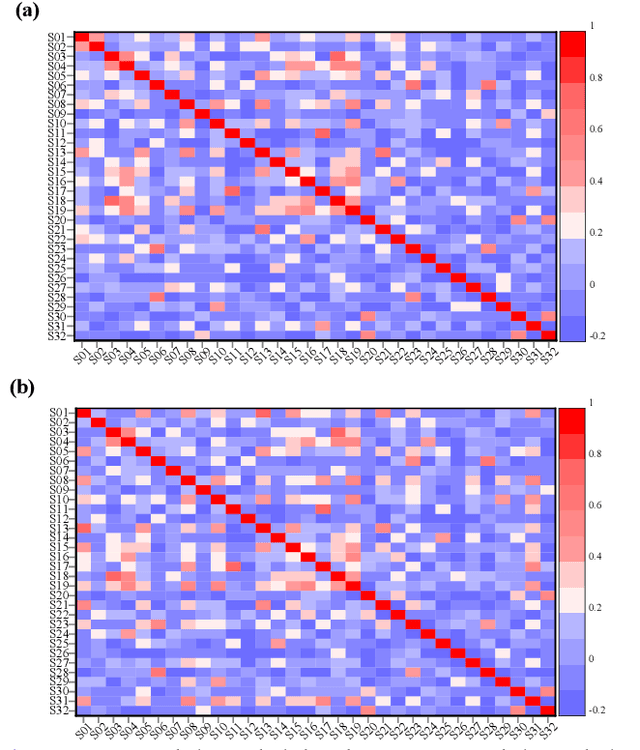

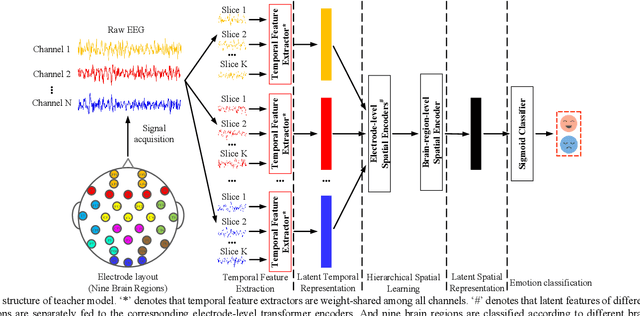

Abstract:Individual differences of Electroencephalogram (EEG) could cause the domain shift which would significantly degrade the performance of cross-subject strategy. The domain adversarial neural networks (DANN), where the classification loss and domain loss jointly update the parameters of feature extractor, are adopted to deal with the domain shift. However, limited EEG data quantity and strong individual difference are challenges for the DANN with cumbersome feature extractor. In this work, we propose knowledge distillation (KD) based lightweight DANN to enhance cross-subject EEG-based emotion recognition. Specifically, the teacher model with strong context learning ability is utilized to learn complex temporal dynamics and spatial correlations of EEG, and robust lightweight student model is guided by the teacher model to learn more difficult domain-invariant features. In the feature-based KD framework, a transformer-based hierarchical temporalspatial learning model is served as the teacher model. The student model, which is composed of Bi-LSTM units, is a lightweight version of the teacher model. Hence, the student model could be supervised to mimic the robust feature representations of teacher model by leveraging complementary latent temporal features and spatial features. In the DANN-based cross-subject emotion recognition, we combine the obtained student model and a lightweight temporal-spatial feature interaction module as the feature extractor. And the feature aggregation is fed to the emotion classifier and domain classifier for domain-invariant feature learning. To verify the effectiveness of the proposed method, we conduct the subject-independent experiments on the public dataset DEAP with arousal and valence classification. The outstanding performance and t-SNE visualization of latent features verify the advantage and effectiveness of the proposed method.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Confident Clustering via PCA Compression Ratio and Its Application to Single-cell RNA-seq Analysis
May 19, 2022Abstract:Unsupervised clustering algorithms for vectors has been widely used in the area of machine learning. Many applications, including the biological data we studied in this paper, contain some boundary datapoints which show combination properties of two underlying clusters and could lower the performance of the traditional clustering algorithms. We develop a confident clustering method aiming to diminish the influence of these datapoints and improve the clustering results. Concretely, for a list of datapoints, we give two clustering results. The first-round clustering attempts to classify only pure vectors with high confidence. Based on it, we classify more vectors with less confidence in the second round. We validate our algorithm on single-cell RNA-seq data, which is a powerful and widely used tool in biology area. Our confident clustering shows a high accuracy on our tested datasets. In addition, unlike traditional clustering methods in single-cell analysis, the confident clustering shows high stability under different choices of parameters.
Compressibility: Power of PCA in Clustering Problems Beyond Dimensionality Reduction
Apr 22, 2022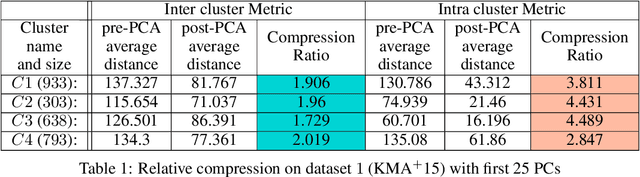
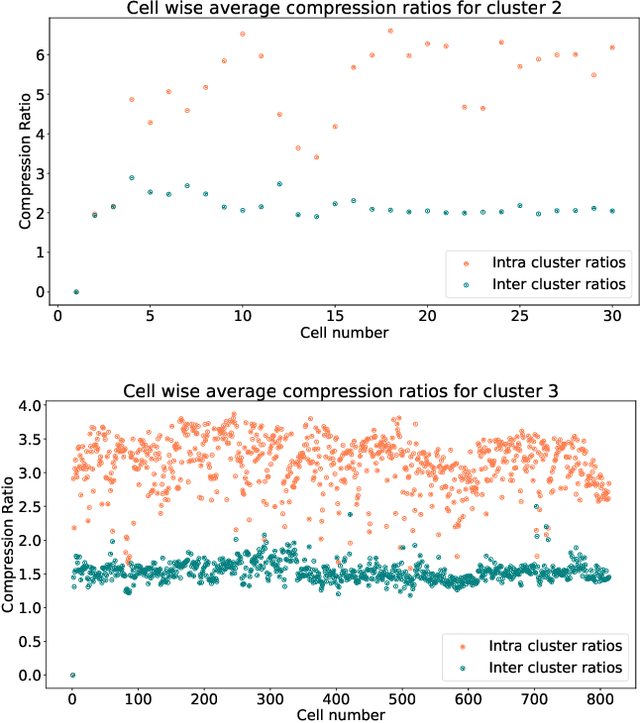
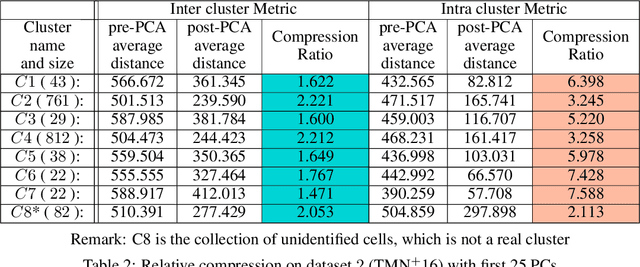
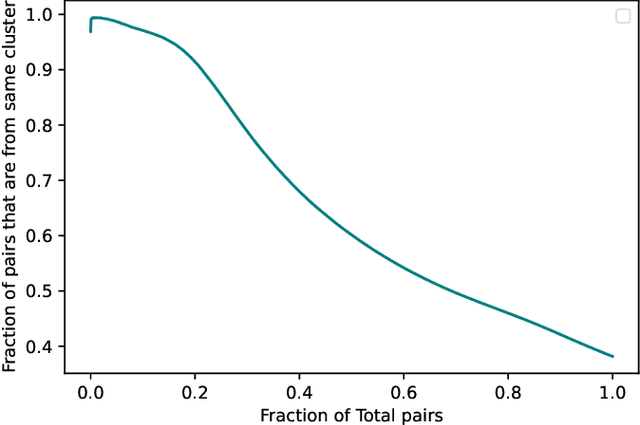
Abstract:In this paper we take a step towards understanding the impact of principle component analysis (PCA) in the context of unsupervised clustering beyond a dimensionality reduction tool. We explore another property of PCA in vector clustering problems, which we call compressibility. This phenomenon shows that PCA significantly reduces the distance of data points belonging to the same clusters, while reducing inter-cluster distances relatively mildly. This gap explains many empirical observations found in practice. For example, in single-cell RNA-sequencing analysis, which is an application of vector clustering in biology, it has been observed that applying PCA on datasets significantly improves the accuracy of classical clustering algorithms such as K-means. We study this compression gap in both theory and practice. On the theoretical side, we analyze PCA in a fairly general probabilistic setup, which we call the random vector model. In terms of practice, we verify the compressibility of PCA on multiple single-cell RNA-seq datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge