Chuan Lei
AgentSM: Semantic Memory for Agentic Text-to-SQL
Jan 22, 2026Abstract:Recent advances in LLM-based Text-to-SQL have achieved remarkable gains on public benchmarks such as BIRD and Spider. Yet, these systems struggle to scale in realistic enterprise settings with large, complex schemas, diverse SQL dialects, and expensive multi-step reasoning. Emerging agentic approaches show potential for adaptive reasoning but often suffer from inefficiency and instability-repeating interactions with databases, producing inconsistent outputs, and occasionally failing to generate valid answers. To address these challenges, we introduce Agent Semantic Memory (AgentSM), an agentic framework for Text-to-SQL that builds and leverages interpretable semantic memory. Instead of relying on raw scratchpads or vector retrieval, AgentSM captures prior execution traces-or synthesizes curated ones-as structured programs that directly guide future reasoning. This design enables systematic reuse of reasoning paths, which allows agents to scale to larger schemas, more complex questions, and longer trajectories efficiently and reliably. Compared to state-of-the-art systems, AgentSM achieves higher efficiency by reducing average token usage and trajectory length by 25% and 35%, respectively, on the Spider 2.0 benchmark. It also improves execution accuracy, reaching a state-of-the-art accuracy of 44.8% on the Spider 2.0 Lite benchmark.
SQLens: An End-to-End Framework for Error Detection and Correction in Text-to-SQL
Jun 04, 2025Abstract:Text-to-SQL systems translate natural language (NL) questions into SQL queries, enabling non-technical users to interact with structured data. While large language models (LLMs) have shown promising results on the text-to-SQL task, they often produce semantically incorrect yet syntactically valid queries, with limited insight into their reliability. We propose SQLens, an end-to-end framework for fine-grained detection and correction of semantic errors in LLM-generated SQL. SQLens integrates error signals from both the underlying database and the LLM to identify potential semantic errors within SQL clauses. It further leverages these signals to guide query correction. Empirical results on two public benchmarks show that SQLens outperforms the best LLM-based self-evaluation method by 25.78% in F1 for error detection, and improves execution accuracy of out-of-the-box text-to-SQL systems by up to 20%.
TailorSQL: An NL2SQL System Tailored to Your Query Workload
May 29, 2025Abstract:NL2SQL (natural language to SQL) translates natural language questions into SQL queries, thereby making structured data accessible to non-technical users, serving as the foundation for intelligent data applications. State-of-the-art NL2SQL techniques typically perform translation by retrieving database-specific information, such as the database schema, and invoking a pre-trained large language model (LLM) using the question and retrieved information to generate the SQL query. However, existing NL2SQL techniques miss a key opportunity which is present in real-world settings: NL2SQL is typically applied on existing databases which have already served many SQL queries in the past. The past query workload implicitly contains information which is helpful for accurate NL2SQL translation and is not apparent from the database schema alone, such as common join paths and the semantics of obscurely-named tables and columns. We introduce TailorSQL, a NL2SQL system that takes advantage of information in the past query workload to improve both the accuracy and latency of translating natural language questions into SQL. By specializing to a given workload, TailorSQL achieves up to 2$\times$ improvement in execution accuracy on standardized benchmarks.
ODIN: A NL2SQL Recommender to Handle Schema Ambiguity
May 25, 2025Abstract:NL2SQL (natural language to SQL) systems translate natural language into SQL queries, allowing users with no technical background to interact with databases and create tools like reports or visualizations. While recent advancements in large language models (LLMs) have significantly improved NL2SQL accuracy, schema ambiguity remains a major challenge in enterprise environments with complex schemas, where multiple tables and columns with semantically similar names often co-exist. To address schema ambiguity, we introduce ODIN, a NL2SQL recommendation engine. Instead of producing a single SQL query given a natural language question, ODIN generates a set of potential SQL queries by accounting for different interpretations of ambiguous schema components. ODIN dynamically adjusts the number of suggestions based on the level of ambiguity, and ODIN learns from user feedback to personalize future SQL query recommendations. Our evaluation shows that ODIN improves the likelihood of generating the correct SQL query by 1.5-2$\times$ compared to baselines.
FeatNavigator: Automatic Feature Augmentation on Tabular Data
Jun 13, 2024Abstract:Data-centric AI focuses on understanding and utilizing high-quality, relevant data in training machine learning (ML) models, thereby increasing the likelihood of producing accurate and useful results. Automatic feature augmentation, aiming to augment the initial base table with useful features from other tables, is critical in data preparation as it improves model performance, robustness, and generalizability. While recent works have investigated automatic feature augmentation, most of them have limited capabilities in utilizing all useful features as many of them are in candidate tables not directly joinable with the base table. Worse yet, with numerous join paths leading to these distant features, existing solutions fail to fully exploit them within a reasonable compute budget. We present FeatNavigator, an effective and efficient framework that explores and integrates high-quality features in relational tables for ML models. FeatNavigator evaluates a feature from two aspects: (1) the intrinsic value of a feature towards an ML task (i.e., feature importance) and (2) the efficacy of a join path connecting the feature to the base table (i.e., integration quality). FeatNavigator strategically selects a small set of available features and their corresponding join paths to train a feature importance estimation model and an integration quality prediction model. Furthermore, FeatNavigator's search algorithm exploits both estimated feature importance and integration quality to identify the optimized feature augmentation plan. Our experimental results show that FeatNavigator outperforms state-of-the-art solutions on five public datasets by up to 40.1% in ML model performance.
4DBInfer: A 4D Benchmarking Toolbox for Graph-Centric Predictive Modeling on Relational DBs
Apr 28, 2024
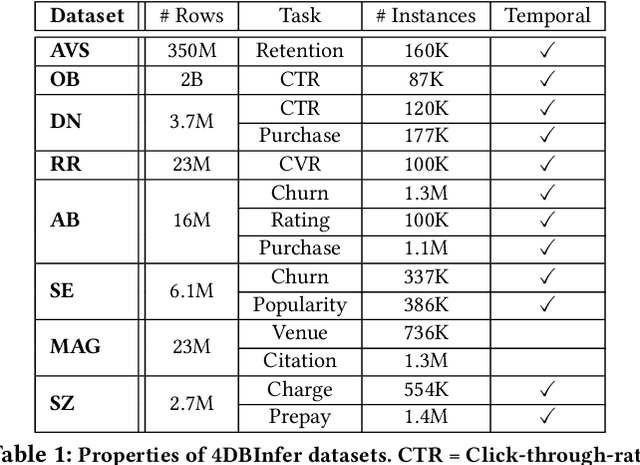
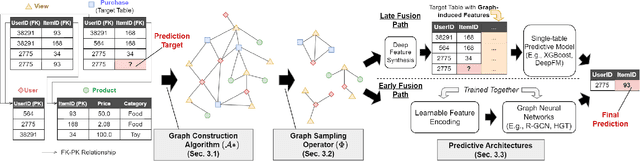
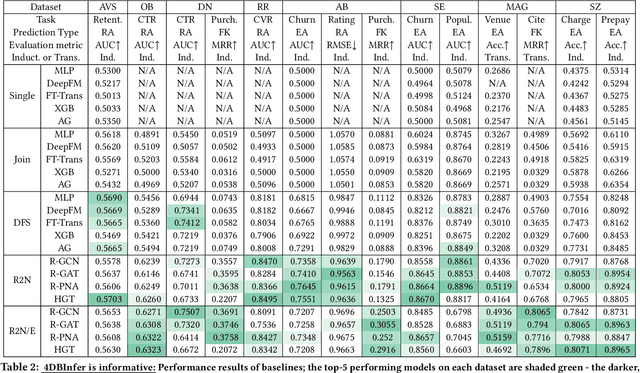
Abstract:Although RDBs store vast amounts of rich, informative data spread across interconnected tables, the progress of predictive machine learning models as applied to such tasks arguably falls well behind advances in other domains such as computer vision or natural language processing. This deficit stems, at least in part, from the lack of established/public RDB benchmarks as needed for training and evaluation purposes. As a result, related model development thus far often defaults to tabular approaches trained on ubiquitous single-table benchmarks, or on the relational side, graph-based alternatives such as GNNs applied to a completely different set of graph datasets devoid of tabular characteristics. To more precisely target RDBs lying at the nexus of these two complementary regimes, we explore a broad class of baseline models predicated on: (i) converting multi-table datasets into graphs using various strategies equipped with efficient subsampling, while preserving tabular characteristics; and (ii) trainable models with well-matched inductive biases that output predictions based on these input subgraphs. Then, to address the dearth of suitable public benchmarks and reduce siloed comparisons, we assemble a diverse collection of (i) large-scale RDB datasets and (ii) coincident predictive tasks. From a delivery standpoint, we operationalize the above four dimensions (4D) of exploration within a unified, scalable open-source toolbox called 4DBInfer. We conclude by presenting evaluations using 4DBInfer, the results of which highlight the importance of considering each such dimension in the design of RDB predictive models, as well as the limitations of more naive approaches such as simply joining adjacent tables. Our source code is released at https://github.com/awslabs/multi-table-benchmark .
OpenTab: Advancing Large Language Models as Open-domain Table Reasoners
Feb 22, 2024



Abstract:Large Language Models (LLMs) trained on large volumes of data excel at various natural language tasks, but they cannot handle tasks requiring knowledge that has not been trained on previously. One solution is to use a retriever that fetches relevant information to expand LLM's knowledge scope. However, existing textual-oriented retrieval-based LLMs are not ideal on structured table data due to diversified data modalities and large table sizes. In this work, we propose OpenTab, an open-domain table reasoning framework powered by LLMs. Overall, OpenTab leverages table retriever to fetch relevant tables and then generates SQL programs to parse the retrieved tables efficiently. Utilizing the intermediate data derived from the SQL executions, it conducts grounded inference to produce accurate response. Extensive experimental evaluation shows that OpenTab significantly outperforms baselines in both open- and closed-domain settings, achieving up to 21.5% higher accuracy. We further run ablation studies to validate the efficacy of our proposed designs of the system.
Scaling Knowledge Graph Embedding Models
Jan 08, 2022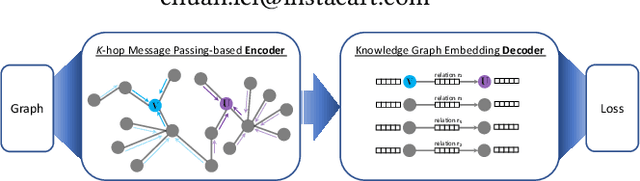
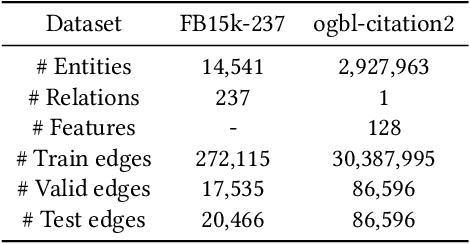
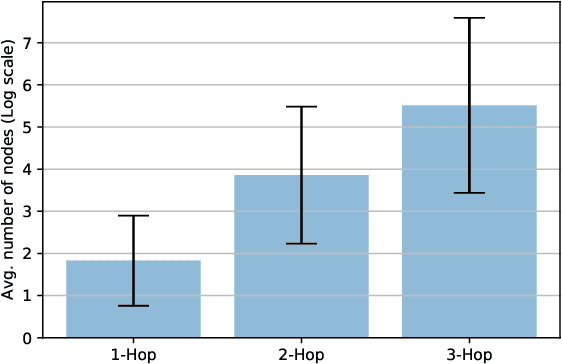
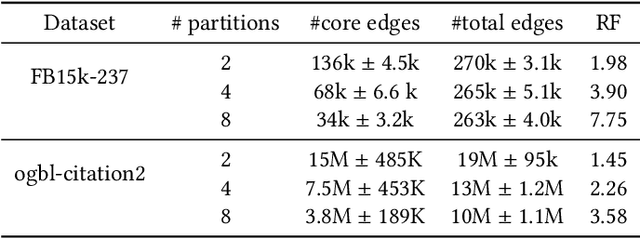
Abstract:Developing scalable solutions for training Graph Neural Networks (GNNs) for link prediction tasks is challenging due to the high data dependencies which entail high computational cost and huge memory footprint. We propose a new method for scaling training of knowledge graph embedding models for link prediction to address these challenges. Towards this end, we propose the following algorithmic strategies: self-sufficient partitions, constraint-based negative sampling, and edge mini-batch training. Both, partitioning strategy and constraint-based negative sampling, avoid cross partition data transfer during training. In our experimental evaluation, we show that our scaling solution for GNN-based knowledge graph embedding models achieves a 16x speed up on benchmark datasets while maintaining a comparable model performance as non-distributed methods on standard metrics.
BI-REC: Guided Data Analysis for Conversational Business Intelligence
May 02, 2021

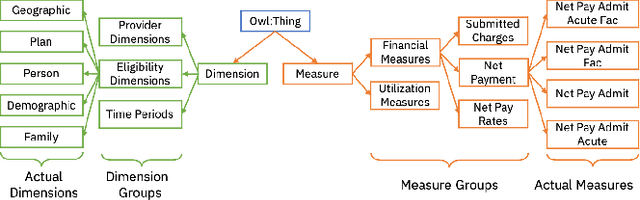
Abstract:Conversational interfaces to Business Intelligence (BI) applications enable data analysis using a natural language dialog in small incremental steps. To truly unleash the power of conversational BI to democratize access to data, a system needs to provide effective and continuous support for data analysis. In this paper, we propose BI-REC, a conversational recommendation system for BI applications to help users accomplish their data analysis tasks. We define the space of data analysis in terms of BI patterns, augmented with rich semantic information extracted from the OLAP cube definition, and use graph embeddings learned using GraphSAGE to create a compact representation of the analysis state. We propose a two-step approach to explore the search space for useful BI pattern recommendations. In the first step, we train a multi-class classifier using prior query logs to predict the next high-level actions in terms of a BI operation (e.g., {\em Drill-Down} or {\em Roll-up}) and a measure that the user is interested in. In the second step, the high-level actions are further refined into actual BI pattern recommendations using collaborative filtering. This two-step approach allows us to not only divide and conquer the huge search space, but also requires less training data. Our experimental evaluation shows that BI-REC achieves an accuracy of 83% for BI pattern recommendations and up to 2X speedup in latency of prediction compared to a state-of-the-art baseline. Our user study further shows that BI-REC provides recommendations with a precision@3 of 91.90% across several different analysis tasks.
Medical Entity Disambiguation Using Graph Neural Networks
Apr 03, 2021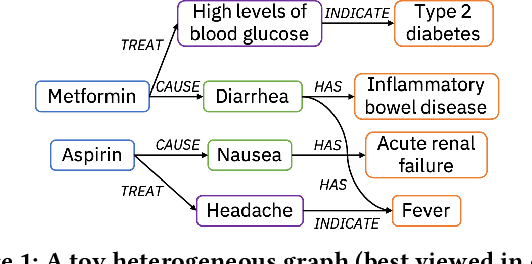
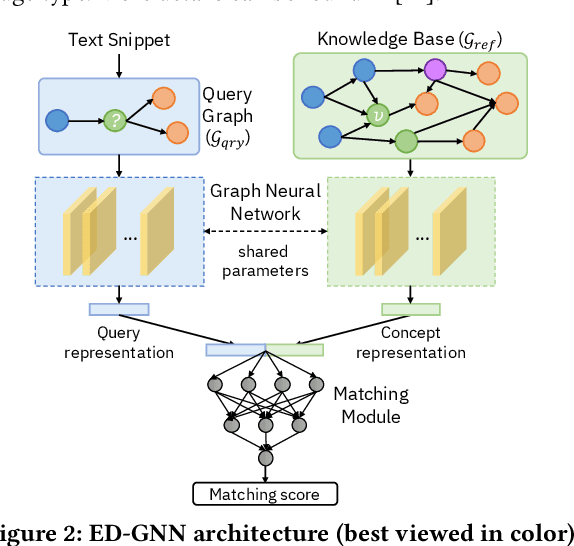
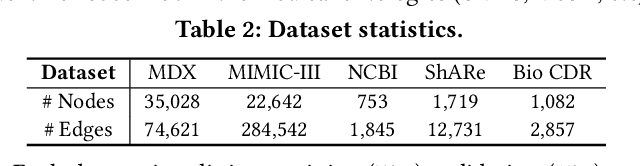
Abstract:Medical knowledge bases (KBs), distilled from biomedical literature and regulatory actions, are expected to provide high-quality information to facilitate clinical decision making. Entity disambiguation (also referred to as entity linking) is considered as an essential task in unlocking the wealth of such medical KBs. However, existing medical entity disambiguation methods are not adequate due to word discrepancies between the entities in the KB and the text snippets in the source documents. Recently, graph neural networks (GNNs) have proven to be very effective and provide state-of-the-art results for many real-world applications with graph-structured data. In this paper, we introduce ED-GNN based on three representative GNNs (GraphSAGE, R-GCN, and MAGNN) for medical entity disambiguation. We develop two optimization techniques to fine-tune and improve ED-GNN. First, we introduce a novel strategy to represent entities that are mentioned in text snippets as a query graph. Second, we design an effective negative sampling strategy that identifies hard negative samples to improve the model's disambiguation capability. Compared to the best performing state-of-the-art solutions, our ED-GNN offers an average improvement of 7.3% in terms of F1 score on five real-world datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge