Fatma Özcan
CardBench: A Benchmark for Learned Cardinality Estimation in Relational Databases
Aug 28, 2024Abstract:Cardinality estimation is crucial for enabling high query performance in relational databases. Recently learned cardinality estimation models have been proposed to improve accuracy but there is no systematic benchmark or datasets which allows researchers to evaluate the progress made by new learned approaches and even systematically develop new learned approaches. In this paper, we are releasing a benchmark, containing thousands of queries over 20 distinct real-world databases for learned cardinality estimation. In contrast to other initial benchmarks, our benchmark is much more diverse and can be used for training and testing learned models systematically. Using this benchmark, we explored whether learned cardinality estimation can be transferred to an unseen dataset in a zero-shot manner. We trained GNN-based and transformer-based models to study the problem in three setups: 1-) instance-based, 2-) zero-shot, and 3-) fine-tuned. Our results show that while we get promising results for zero-shot cardinality estimation on simple single table queries; as soon as we add joins, the accuracy drops. However, we show that with fine-tuning, we can still utilize pre-trained models for cardinality estimation, significantly reducing training overheads compared to instance specific models. We are open sourcing our scripts to collect statistics, generate queries and training datasets to foster more extensive research, also from the ML community on the important problem of cardinality estimation and in particular improve on recent directions such as pre-trained cardinality estimation.
Medical Entity Disambiguation Using Graph Neural Networks
Apr 03, 2021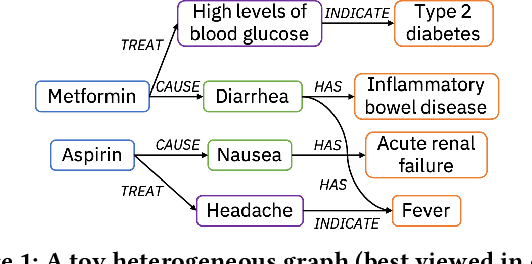
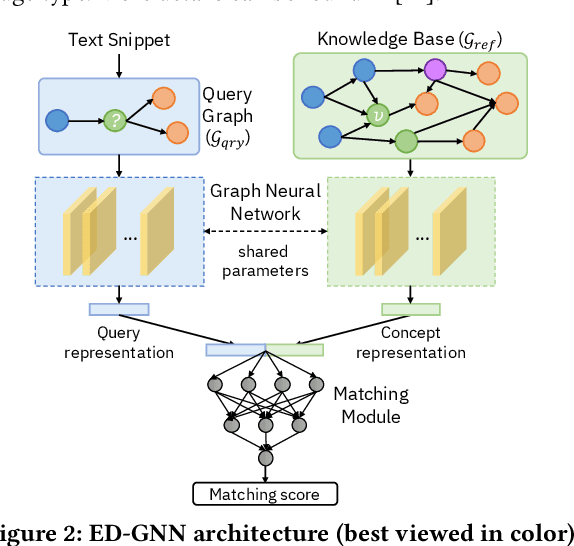
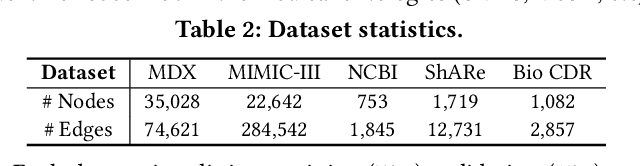
Abstract:Medical knowledge bases (KBs), distilled from biomedical literature and regulatory actions, are expected to provide high-quality information to facilitate clinical decision making. Entity disambiguation (also referred to as entity linking) is considered as an essential task in unlocking the wealth of such medical KBs. However, existing medical entity disambiguation methods are not adequate due to word discrepancies between the entities in the KB and the text snippets in the source documents. Recently, graph neural networks (GNNs) have proven to be very effective and provide state-of-the-art results for many real-world applications with graph-structured data. In this paper, we introduce ED-GNN based on three representative GNNs (GraphSAGE, R-GCN, and MAGNN) for medical entity disambiguation. We develop two optimization techniques to fine-tune and improve ED-GNN. First, we introduce a novel strategy to represent entities that are mentioned in text snippets as a query graph. Second, we design an effective negative sampling strategy that identifies hard negative samples to improve the model's disambiguation capability. Compared to the best performing state-of-the-art solutions, our ED-GNN offers an average improvement of 7.3% in terms of F1 score on five real-world datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge