Chengbin Hou
Enhancing Noise Robustness of Parkinson's Disease Telemonitoring via Contrastive Feature Augmentation
Oct 02, 2025Abstract:Parkinson's disease (PD) is one of the most common neurodegenerative disorder. PD telemonitoring emerges as a novel assessment modality enabling self-administered at-home tests of Unified Parkinson's Disease Rating Scale (UPDRS) scores, enhancing accessibility for PD patients. However, three types of noise would occur during measurements: (1) patient-induced measurement inaccuracies, (2) environmental noise, and (3) data packet loss during transmission, resulting in higher prediction errors. To address these challenges, NoRo, a noise-robust UPDRS prediction framework is proposed. First, the original speech features are grouped into ordered bins, based on the continuous values of a selected feature, to construct contrastive pairs. Second, the contrastive pairs are employed to train a multilayer perceptron encoder for generating noise-robust features. Finally, these features are concatenated with the original features as the augmented features, which are then fed into the UPDRS prediction models. Notably, we further introduces a novel evaluation approach with customizable noise injection module, and extensive experiments show that NoRo can successfully enhance the noise robustness of UPDRS prediction across various downstream prediction models under different noisy environments.
Look Inside for More: Internal Spatial Modality Perception for 3D Anomaly Detection
Dec 18, 2024



Abstract:3D anomaly detection has recently become a significant focus in computer vision. Several advanced methods have achieved satisfying anomaly detection performance. However, they typically concentrate on the external structure of 3D samples and struggle to leverage the internal information embedded within samples. Inspired by the basic intuition of why not look inside for more, we introduce a straightforward method named Internal Spatial Modality Perception (ISMP) to explore the feature representation from internal views fully. Specifically, our proposed ISMP consists of a critical perception module, Spatial Insight Engine (SIE), which abstracts complex internal information of point clouds into essential global features. Besides, to better align structural information with point data, we propose an enhanced key point feature extraction module for amplifying spatial structure feature representation. Simultaneously, a novel feature filtering module is incorporated to reduce noise and redundant features for further aligning precise spatial structure. Extensive experiments validate the effectiveness of our proposed method, achieving object-level and pixel-level AUROC improvements of 4.2% and 13.1%, respectively, on the Real3D-AD benchmarks. Note that the strong generalization ability of SIE has been theoretically proven and is verified in both classification and segmentation tasks.
Node Importance Estimation Leveraging LLMs for Semantic Augmentation in Knowledge Graphs
Nov 30, 2024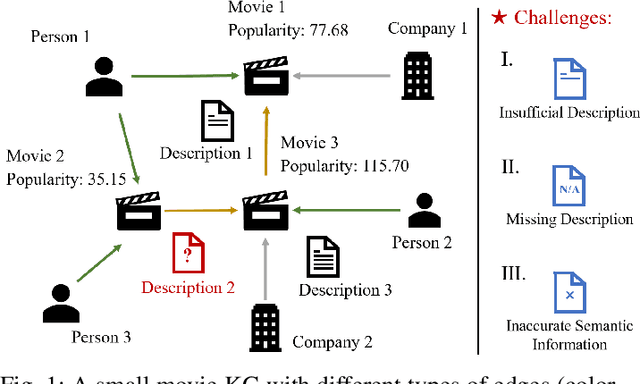
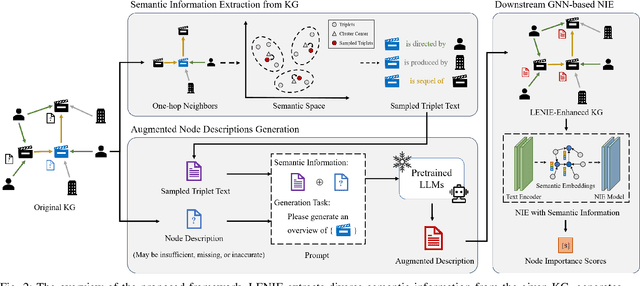


Abstract:Node Importance Estimation (NIE) is a task that quantifies the importance of node in a graph. Recent research has investigated to exploit various information from Knowledge Graphs (KGs) to estimate node importance scores. However, the semantic information in KGs could be insufficient, missing, and inaccurate, which would limit the performance of existing NIE models. To address these issues, we leverage Large Language Models (LLMs) for semantic augmentation thanks to the LLMs' extra knowledge and ability of integrating knowledge from both LLMs and KGs. To this end, we propose the LLMs Empowered Node Importance Estimation (LENIE) method to enhance the semantic information in KGs for better supporting NIE tasks. To our best knowledge, this is the first work incorporating LLMs into NIE. Specifically, LENIE employs a novel clustering-based triplet sampling strategy to extract diverse knowledge of a node sampled from the given KG. After that, LENIE adopts the node-specific adaptive prompts to integrate the sampled triplets and the original node descriptions, which are then fed into LLMs for generating richer and more precise augmented node descriptions. These augmented descriptions finally initialize node embeddings for boosting the downstream NIE model performance. Extensive experiments demonstrate LENIE's effectiveness in addressing semantic deficiencies in KGs, enabling more informative semantic augmentation and enhancing existing NIE models to achieve the state-of-the-art performance. The source code of LENIE is freely available at \url{https://github.com/XinyuLin-FZ/LENIE}.
Molecular Graph Representation Learning Integrating Large Language Models with Domain-specific Small Models
Aug 19, 2024



Abstract:Molecular property prediction is a crucial foundation for drug discovery. In recent years, pre-trained deep learning models have been widely applied to this task. Some approaches that incorporate prior biological domain knowledge into the pre-training framework have achieved impressive results. However, these methods heavily rely on biochemical experts, and retrieving and summarizing vast amounts of domain knowledge literature is both time-consuming and expensive. Large Language Models (LLMs) have demonstrated remarkable performance in understanding and efficiently providing general knowledge. Nevertheless, they occasionally exhibit hallucinations and lack precision in generating domain-specific knowledge. Conversely, Domain-specific Small Models (DSMs) possess rich domain knowledge and can accurately calculate molecular domain-related metrics. However, due to their limited model size and singular functionality, they lack the breadth of knowledge necessary for comprehensive representation learning. To leverage the advantages of both approaches in molecular property prediction, we propose a novel Molecular Graph representation learning framework that integrates Large language models and Domain-specific small models (MolGraph-LarDo). Technically, we design a two-stage prompt strategy where DSMs are introduced to calibrate the knowledge provided by LLMs, enhancing the accuracy of domain-specific information and thus enabling LLMs to generate more precise textual descriptions for molecular samples. Subsequently, we employ a multi-modal alignment method to coordinate various modalities, including molecular graphs and their corresponding descriptive texts, to guide the pre-training of molecular representations. Extensive experiments demonstrate the effectiveness of the proposed method.
Towards High-resolution 3D Anomaly Detection via Group-Level Feature Contrastive Learning
Aug 08, 2024



Abstract:High-resolution point clouds~(HRPCD) anomaly detection~(AD) plays a critical role in precision machining and high-end equipment manufacturing. Despite considerable 3D-AD methods that have been proposed recently, they still cannot meet the requirements of the HRPCD-AD task. There are several challenges: i) It is difficult to directly capture HRPCD information due to large amounts of points at the sample level; ii) The advanced transformer-based methods usually obtain anisotropic features, leading to degradation of the representation; iii) The proportion of abnormal areas is very small, which makes it difficult to characterize. To address these challenges, we propose a novel group-level feature-based network, called Group3AD, which has a significantly efficient representation ability. First, we design an Intercluster Uniformity Network~(IUN) to present the mapping of different groups in the feature space as several clusters, and obtain a more uniform distribution between clusters representing different parts of the point clouds in the feature space. Then, an Intracluster Alignment Network~(IAN) is designed to encourage groups within the cluster to be distributed tightly in the feature space. In addition, we propose an Adaptive Group-Center Selection~(AGCS) based on geometric information to improve the pixel density of potential anomalous regions during inference. The experimental results verify the effectiveness of our proposed Group3AD, which surpasses Reg3D-AD by the margin of 5\% in terms of object-level AUROC on Real3D-AD. We provide the code and supplementary information on our website: https://github.com/M-3LAB/Group3AD.
Label Informed Contrastive Pretraining for Node Importance Estimation on Knowledge Graphs
Feb 26, 2024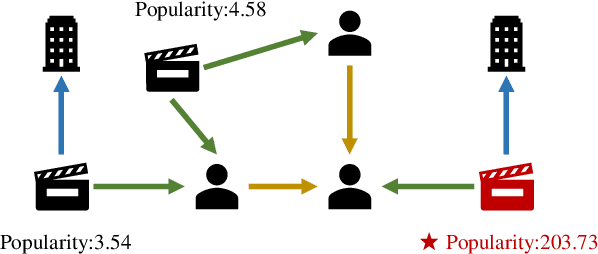
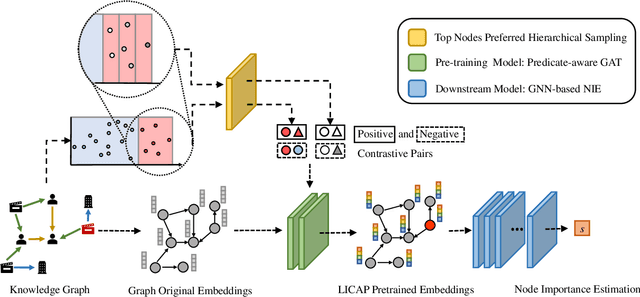
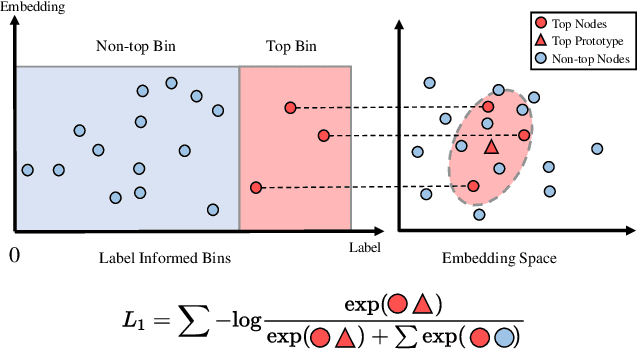
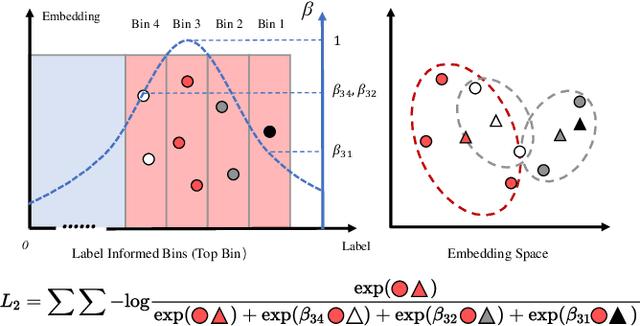
Abstract:Node Importance Estimation (NIE) is a task of inferring importance scores of the nodes in a graph. Due to the availability of richer data and knowledge, recent research interests of NIE have been dedicating to knowledge graphs for predicting future or missing node importance scores. Existing state-of-the-art NIE methods train the model by available labels, and they consider every interested node equally before training. However, the nodes with higher importance often require or receive more attention in real-world scenarios, e.g., people may care more about the movies or webpages with higher importance. To this end, we introduce Label Informed ContrAstive Pretraining (LICAP) to the NIE problem for being better aware of the nodes with high importance scores. Specifically, LICAP is a novel type of contrastive learning framework that aims to fully utilize the continuous labels to generate contrastive samples for pretraining embeddings. Considering the NIE problem, LICAP adopts a novel sampling strategy called top nodes preferred hierarchical sampling to first group all interested nodes into a top bin and a non-top bin based on node importance scores, and then divide the nodes within top bin into several finer bins also based on the scores. The contrastive samples are generated from those bins, and are then used to pretrain node embeddings of knowledge graphs via a newly proposed Predicate-aware Graph Attention Networks (PreGAT), so as to better separate the top nodes from non-top nodes, and distinguish the top nodes within top bin by keeping the relative order among finer bins. Extensive experiments demonstrate that the LICAP pretrained embeddings can further boost the performance of existing NIE methods and achieve the new state-of-the-art performance regarding both regression and ranking metrics. The source code for reproducibility is available at https://github.com/zhangtia16/LICAP
Fossil Image Identification using Deep Learning Ensembles of Data Augmented Multiviews
Feb 16, 2023Abstract:Identification of fossil species is crucial to evolutionary studies. Recent advances from deep learning have shown promising prospects in fossil image identification. However, the quantity and quality of labeled fossil images are often limited due to fossil preservation, conditioned sampling, and expensive and inconsistent label annotation by domain experts, which pose great challenges to the training of deep learning based image classification models. To address these challenges, we follow the idea of the wisdom of crowds and propose a novel multiview ensemble framework, which collects multiple views of each fossil specimen image reflecting its different characteristics to train multiple base deep learning models and then makes final decisions via soft voting. We further develop OGS method that integrates original, gray, and skeleton views under this framework to demonstrate the effectiveness. Experimental results on the fusulinid fossil dataset over five deep learning based milestone models show that OGS using three base models consistently outperforms the baseline using a single base model, and the ablation study verifies the usefulness of each selected view. Besides, OGS obtains the superior or comparable performance compared to the method under well-known bagging framework. Moreover, as the available training data decreases, the proposed framework achieves more performance gains compared to the baseline. Furthermore, a consistency test with two human experts shows that OGS obtains the highest agreement with both the labels of dataset and the two experts. Notably, this methodology is designed for general fossil identification and it is expected to see applications on other fossil datasets. The results suggest the potential application when the quantity and quality of labeled data are particularly restricted, e.g., to identify rare fossil images.
A Survey of Trustworthy Graph Learning: Reliability, Explainability, and Privacy Protection
May 23, 2022



Abstract:Deep graph learning has achieved remarkable progresses in both business and scientific areas ranging from finance and e-commerce, to drug and advanced material discovery. Despite these progresses, how to ensure various deep graph learning algorithms behave in a socially responsible manner and meet regulatory compliance requirements becomes an emerging problem, especially in risk-sensitive domains. Trustworthy graph learning (TwGL) aims to solve the above problems from a technical viewpoint. In contrast to conventional graph learning research which mainly cares about model performance, TwGL considers various reliability and safety aspects of the graph learning framework including but not limited to robustness, explainability, and privacy. In this survey, we provide a comprehensive review of recent leading approaches in the TwGL field from three dimensions, namely, reliability, explainability, and privacy protection. We give a general categorization for existing work and review typical work for each category. To give further insights for TwGL research, we provide a unified view to inspect previous works and build the connection between them. We also point out some important open problems remaining to be solved in the future developments of TwGL.
Recent Advances in Reliable Deep Graph Learning: Adversarial Attack, Inherent Noise, and Distribution Shift
Feb 15, 2022


Abstract:Deep graph learning (DGL) has achieved remarkable progress in both business and scientific areas ranging from finance and e-commerce to drug and advanced material discovery. Despite the progress, applying DGL to real-world applications faces a series of reliability threats including adversarial attacks, inherent noise, and distribution shift. This survey aims to provide a comprehensive review of recent advances for improving the reliability of DGL algorithms against the above threats. In contrast to prior related surveys which mainly focus on adversarial attacks and defense, our survey covers more reliability-related aspects of DGL, i.e., inherent noise and distribution shift. Additionally, we discuss the relationships among above aspects and highlight some important issues to be explored in future research.
Robust Dynamic Network Embedding via Ensembles
May 30, 2021



Abstract:Dynamic Network Embedding (DNE) has recently attracted considerable attention due to the advantage of network embedding in various applications and the dynamic nature of many real-world networks. For dynamic networks, the degree of changes, i.e., defined as the averaged number of changed edges between consecutive snapshots spanning a dynamic network, could be very different in real-world scenarios. Although quite a few DNE methods have been proposed, it still remains unclear that whether and to what extent the existing DNE methods are robust to the degree of changes, which is however an important factor in both academic research and industrial applications. In this work, we investigate the robustness issue of DNE methods w.r.t. the degree of changes for the first time and accordingly, propose a robust DNE method. Specifically, the proposed method follows the notion of ensembles where the base learner adopts an incremental Skip-Gram neural embedding approach. To further boost the performance, a novel strategy is proposed to enhance the diversity among base learners at each timestep by capturing different levels of local-global topology. Extensive experiments demonstrate the benefits of special designs in the proposed method, and the superior performance of the proposed method compared to state-of-the-art methods. The comparative study also reveals the robustness issue of some DNE methods. The source code is available at https://github.com/houchengbin/SG-EDNE
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge