Charles Tapley Hoyt
The Cell Ontology in the age of single-cell omics
Jun 10, 2025Abstract:Single-cell omics technologies have transformed our understanding of cellular diversity by enabling high-resolution profiling of individual cells. However, the unprecedented scale and heterogeneity of these datasets demand robust frameworks for data integration and annotation. The Cell Ontology (CL) has emerged as a pivotal resource for achieving FAIR (Findable, Accessible, Interoperable, and Reusable) data principles by providing standardized, species-agnostic terms for canonical cell types - forming a core component of a wide range of platforms and tools. In this paper, we describe the wide variety of uses of CL in these platforms and tools and detail ongoing work to improve and extend CL content including the addition of transcriptomically defined types, working closely with major atlasing efforts including the Human Cell Atlas and the Brain Initiative Cell Atlas Network to support their needs. We cover the challenges and future plans for harmonising classical and transcriptomic cell type definitions, integrating markers and using Large Language Models (LLMs) to improve content and efficiency of CL workflows.
An Open-Source Knowledge Graph Ecosystem for the Life Sciences
Jul 11, 2023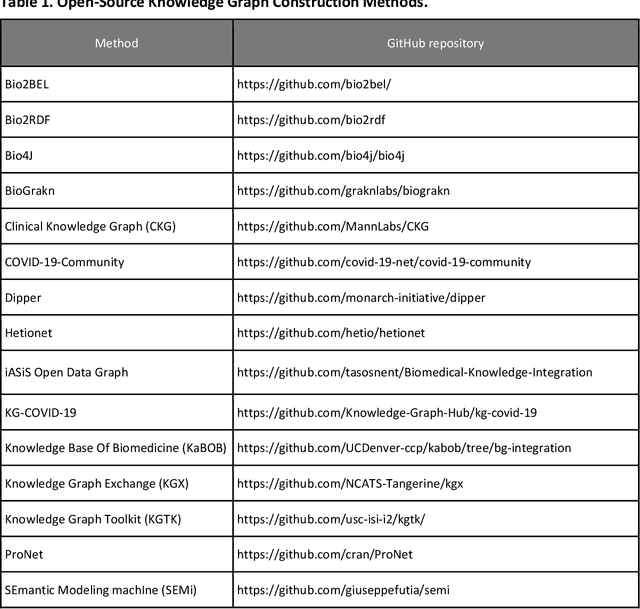
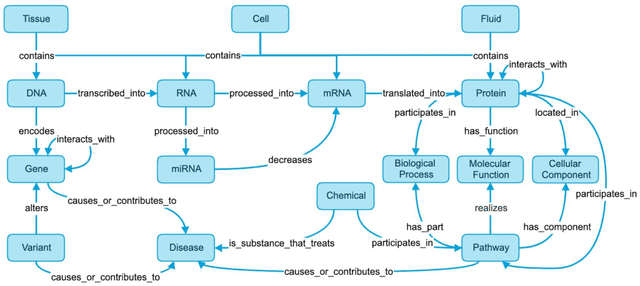
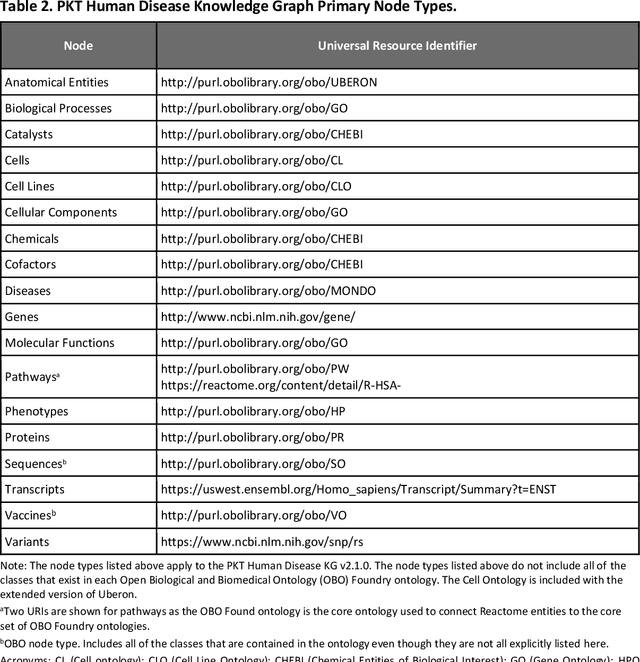
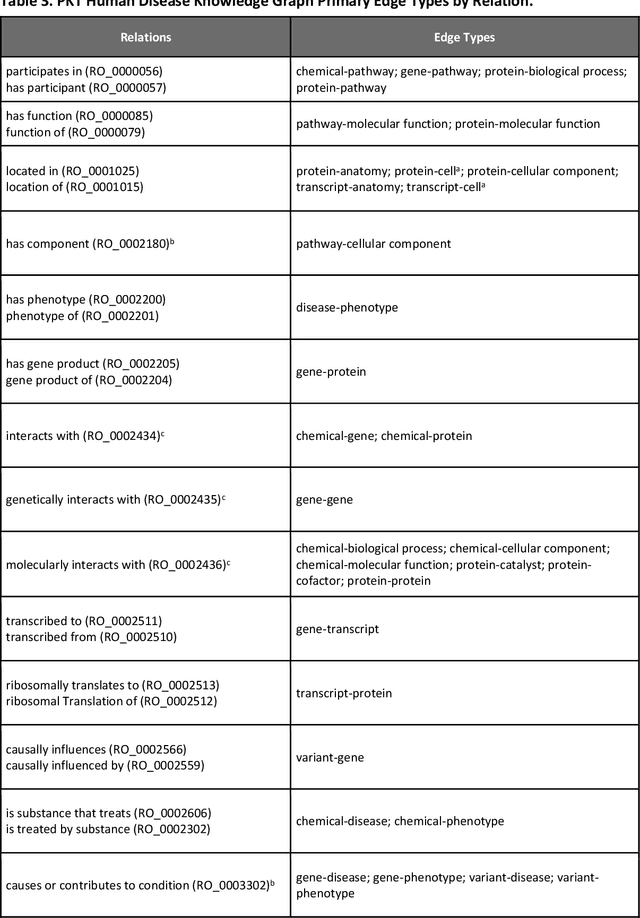
Abstract:Translational research requires data at multiple scales of biological organization. Advancements in sequencing and multi-omics technologies have increased the availability of these data but researchers face significant integration challenges. Knowledge graphs (KGs) are used to model complex phenomena, and methods exist to automatically construct them. However, tackling complex biomedical integration problems requires flexibility in the way knowledge is modeled. Moreover, existing KG construction methods provide robust tooling at the cost of fixed or limited choices among knowledge representation models. PheKnowLator (Phenotype Knowledge Translator) is a semantic ecosystem for automating the FAIR (Findable, Accessible, Interoperable, and Reusable) construction of ontologically grounded KGs with fully customizable knowledge representation. The ecosystem includes KG construction resources (e.g., data preparation APIs), analysis tools (e.g., SPARQL endpoints and abstraction algorithms), and benchmarks (e.g., prebuilt KGs and embeddings). We evaluate the ecosystem by surveying open-source KG construction methods and analyzing its computational performance when constructing 12 large-scale KGs. With flexible knowledge representation, PheKnowLator enables fully customizable KGs without compromising performance or usability.
A Unified Framework for Rank-based Evaluation Metrics for Link Prediction in Knowledge Graphs
Mar 14, 2022



Abstract:The link prediction task on knowledge graphs without explicit negative triples in the training data motivates the usage of rank-based metrics. Here, we review existing rank-based metrics and propose desiderata for improved metrics to address lack of interpretability and comparability of existing metrics to datasets of different sizes and properties. We introduce a simple theoretical framework for rank-based metrics upon which we investigate two avenues for improvements to existing metrics via alternative aggregation functions and concepts from probability theory. We finally propose several new rank-based metrics that are more easily interpreted and compared accompanied by a demonstration of their usage in a benchmarking of knowledge graph embedding models.
An Open Challenge for Inductive Link Prediction on Knowledge Graphs
Mar 03, 2022



Abstract:An emerging trend in representation learning over knowledge graphs (KGs) moves beyond transductive link prediction tasks over a fixed set of known entities in favor of inductive tasks that imply training on one graph and performing inference over a new graph with unseen entities. In inductive setups, node features are often not available and training shallow entity embedding matrices is meaningless as they cannot be used at inference time with unseen entities. Despite the growing interest, there are not enough benchmarks for evaluating inductive representation learning methods. In this work, we introduce ILPC 2022, a novel open challenge on KG inductive link prediction. To this end, we constructed two new datasets based on Wikidata with various sizes of training and inference graphs that are much larger than existing inductive benchmarks. We also provide two strong baselines leveraging recently proposed inductive methods. We hope this challenge helps to streamline community efforts in the inductive graph representation learning area. ILPC 2022 follows best practices on evaluation fairness and reproducibility, and is available at https://github.com/pykeen/ilpc2022.
ChemicalX: A Deep Learning Library for Drug Pair Scoring
Feb 14, 2022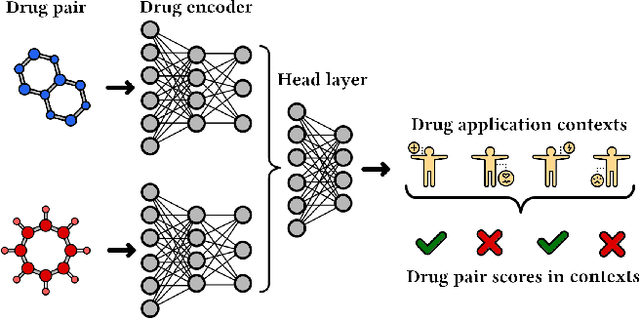
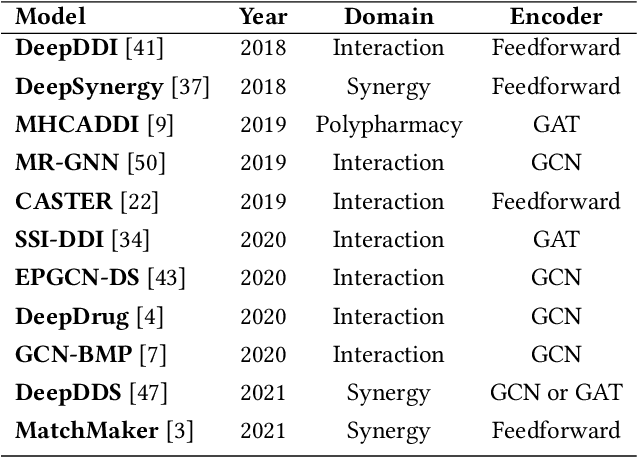
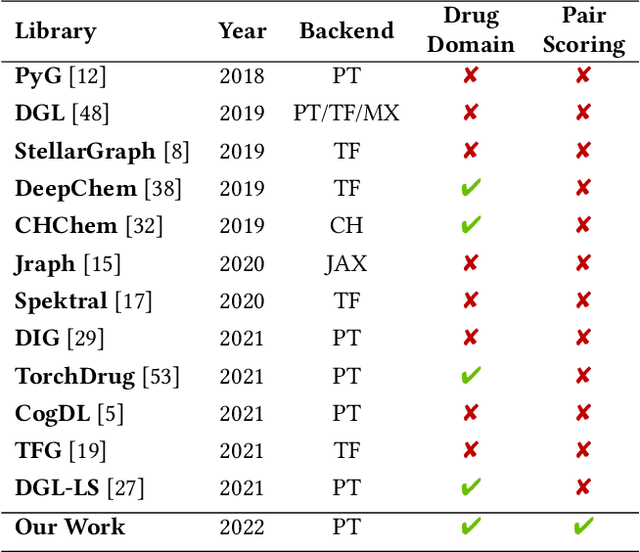
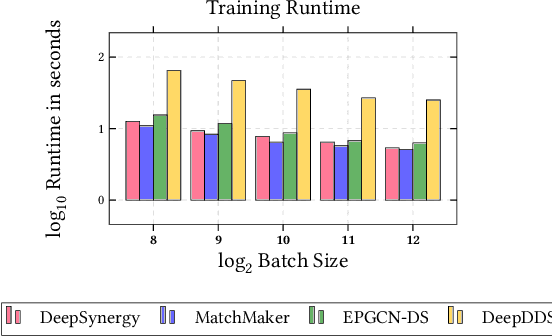
Abstract:In this paper, we introduce ChemicalX, a PyTorch-based deep learning library designed for providing a range of state of the art models to solve the drug pair scoring task. The primary objective of the library is to make deep drug pair scoring models accessible to machine learning researchers and practitioners in a streamlined framework.The design of ChemicalX reuses existing high level model training utilities, geometric deep learning, and deep chemistry layers from the PyTorch ecosystem. Our system provides neural network layers, custom pair scoring architectures, data loaders, and batch iterators for end users. We showcase these features with example code snippets and case studies to highlight the characteristics of ChemicalX. A range of experiments on real world drug-drug interaction, polypharmacy side effect, and combination synergy prediction tasks demonstrate that the models available in ChemicalX are effective at solving the pair scoring task. Finally, we show that ChemicalX could be used to train and score machine learning models on large drug pair datasets with hundreds of thousands of compounds on commodity hardware.
Wavelet-Packet Powered Deepfake Image Detection
Jun 17, 2021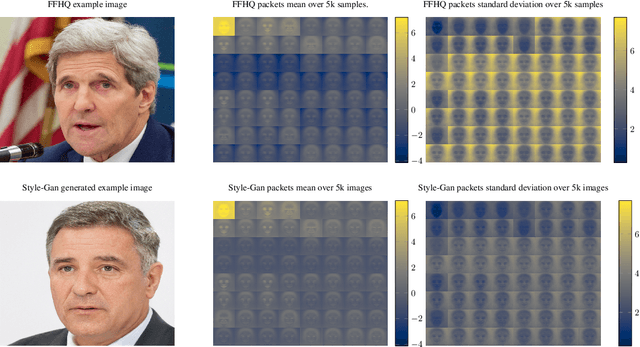
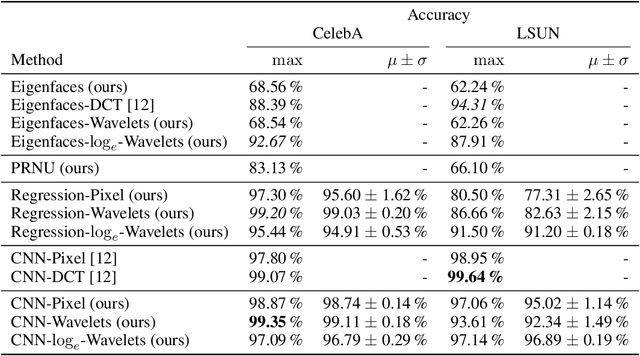

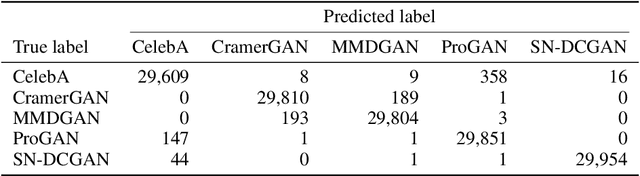
Abstract:As neural networks become more able to generate realistic artificial images, they have the potential to improve movies, music, video games and make the internet an even more creative and inspiring place. Yet, at the same time, the latest technology potentially enables new digital ways to lie. In response, the need for a diverse and reliable toolbox arises to identify artificial images and other content. Previous work primarily relies on pixel-space CNN or the Fourier transform. To the best of our knowledge, wavelet-based gan analysis and detection methods have been absent thus far. This paper aims to fill this gap and describes a wavelet-based approach to gan-generated image analysis and detection. We evaluate our method on FFHQ, CelebA, and LSUN source identification problems and find improved or competitive performance.
Understanding the Performance of Knowledge Graph Embeddings in Drug Discovery
Jun 07, 2021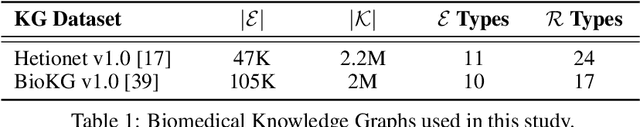
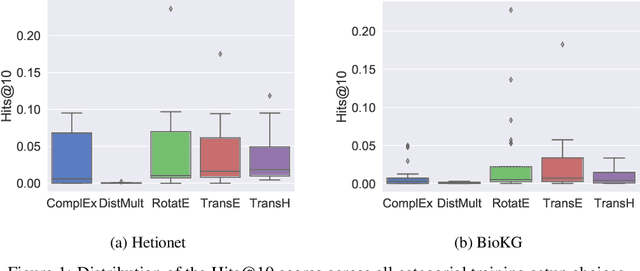
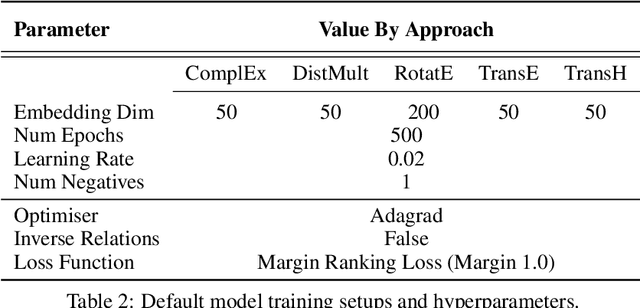
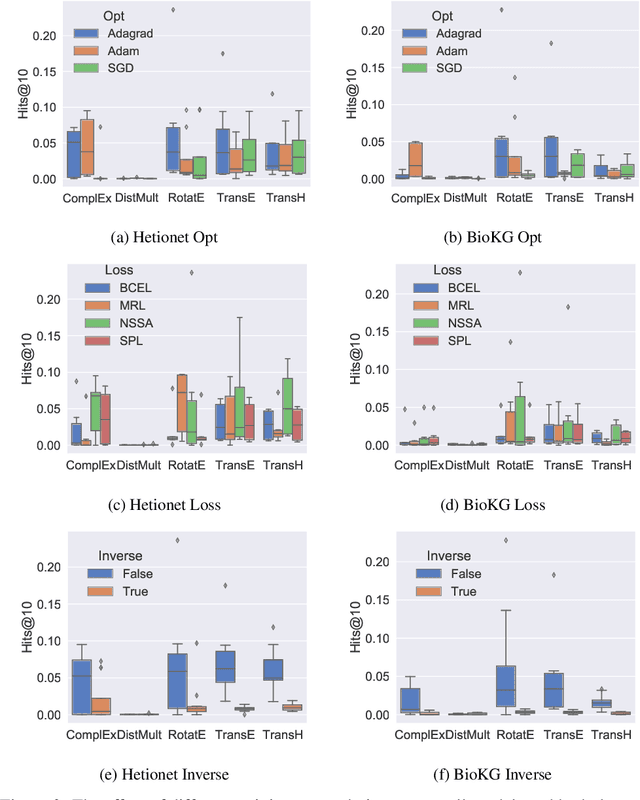
Abstract:Knowledge Graphs (KG) and associated Knowledge Graph Embedding (KGE) models have recently begun to be explored in the context of drug discovery and have the potential to assist in key challenges such as target identification. In the drug discovery domain, KGs can be employed as part of a process which can result in lab-based experiments being performed, or impact on other decisions, incurring significant time and financial costs and most importantly, ultimately influencing patient healthcare. For KGE models to have impact in this domain, a better understanding of not only of performance, but also the various factors which determine it, is required. In this study we investigate, over the course of many thousands of experiments, the predictive performance of five KGE models on two public drug discovery-oriented KGs. Our goal is not to focus on the best overall model or configuration, instead we take a deeper look at how performance can be affected by changes in the training setup, choice of hyperparameters, model parameter initialisation seed and different splits of the datasets. Our results highlight that these factors have significant impact on performance and can even affect the ranking of models. Indeed these factors should be reported along with model architectures to ensure complete reproducibility and fair comparisons of future work, and we argue this is critical for the acceptance of use, and impact of KGEs in a biomedical setting. To aid reproducibility of our own work, we release all experimentation code.
Leveraging Structured Biological Knowledge for Counterfactual Inference: a Case Study of Viral Pathogenesis
Jan 13, 2021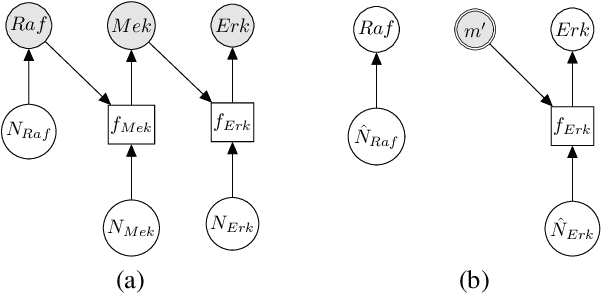
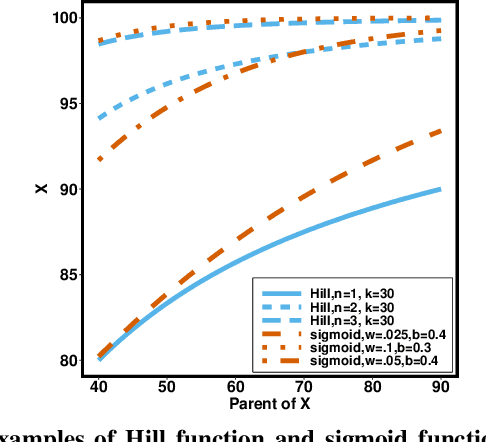
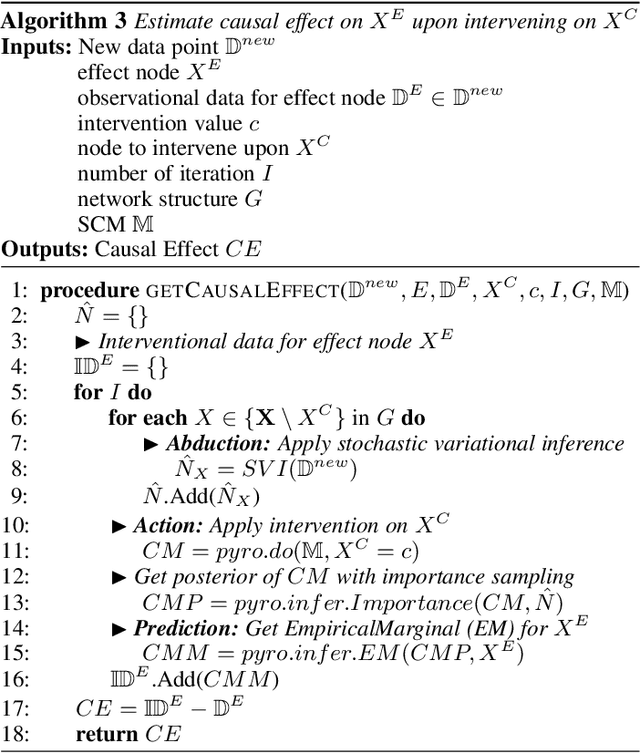
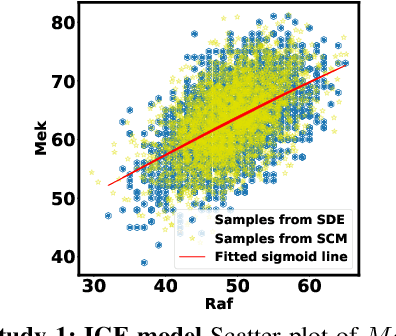
Abstract:Counterfactual inference is a useful tool for comparing outcomes of interventions on complex systems. It requires us to represent the system in form of a structural causal model, complete with a causal diagram, probabilistic assumptions on exogenous variables, and functional assignments. Specifying such models can be extremely difficult in practice. The process requires substantial domain expertise, and does not scale easily to large systems, multiple systems, or novel system modifications. At the same time, many application domains, such as molecular biology, are rich in structured causal knowledge that is qualitative in nature. This manuscript proposes a general approach for querying a causal biological knowledge graph, and converting the qualitative result into a quantitative structural causal model that can learn from data to answer the question. We demonstrate the feasibility, accuracy and versatility of this approach using two case studies in systems biology. The first demonstrates the appropriateness of the underlying assumptions and the accuracy of the results. The second demonstrates the versatility of the approach by querying a knowledge base for the molecular determinants of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced cytokine storm, and performing counterfactual inference to estimate the causal effect of medical countermeasures for severely ill patients.
PyKEEN 1.0: A Python Library for Training and Evaluating Knowledge Graph Embeddings
Jul 30, 2020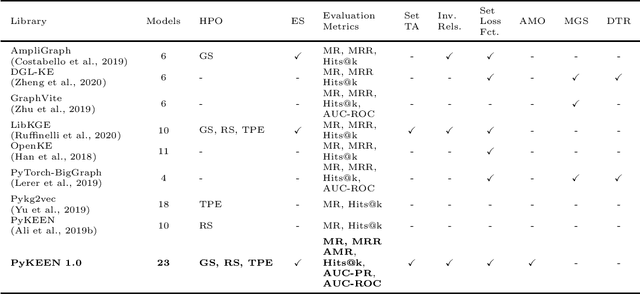
Abstract:Recently, knowledge graph embeddings (KGEs) received significant attention, and several software libraries have been developed for training and evaluating KGEs. While each of them addresses specific needs, we re-designed and re-implemented PyKEEN, one of the first KGE libraries, in a community effort. PyKEEN 1.0 enables users to compose knowledge graph embedding models (KGEMs) based on a wide range of interaction models, training approaches, loss functions, and permits the explicit modeling of inverse relations. Besides, an automatic memory optimization has been realized in order to exploit the provided hardware optimally, and through the integration of Optuna extensive hyper-parameter optimization (HPO) functionalities are provided.
Bringing Light Into the Dark: A Large-scale Evaluation of Knowledge Graph Embedding Models Under a Unified Framework
Jun 23, 2020


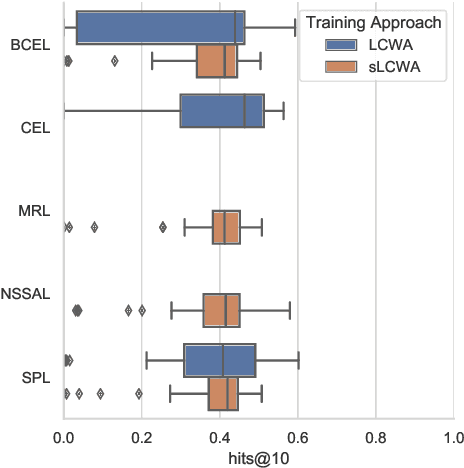
Abstract:The heterogeneity in recently published knowledge graph embedding models' implementations, training, and evaluation has made fair and thorough comparisons difficult. In order to assess the reproducibility of previously published results, we re-implemented and evaluated 19 interaction models in the PyKEEN software package. Here, we outline which results could be reproduced with their reported hyper-parameters, which could only be reproduced with alternate hyper-parameters, and which could not be reproduced at all as well as provide insight as to why this might be the case. We then performed a large-scale benchmarking on four datasets with several thousands of experiments and 21,246 GPU hours of computation time. We present insights gained as to best practices, best configurations for each model, and where improvements could be made over previously published best configurations. Our results highlight that the combination of model architecture, training approach, loss function, and the explicit modeling of inverse relations is crucial for a model's performances, and not only determined by the model architecture. We provide evidence that several architectures can obtain results competitive to the state-of-the-art when configured carefully. We have made all code, experimental configurations, results, and analyses that lead to our interpretations available at https://github.com/pykeen/pykeen and https://github.com/pykeen/benchmarking
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge