Sebastian Nilsson
ChemicalX: A Deep Learning Library for Drug Pair Scoring
Feb 14, 2022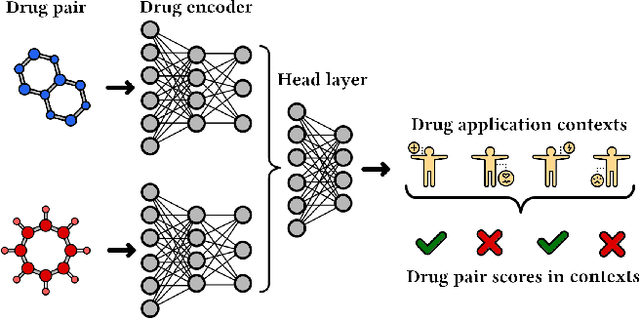
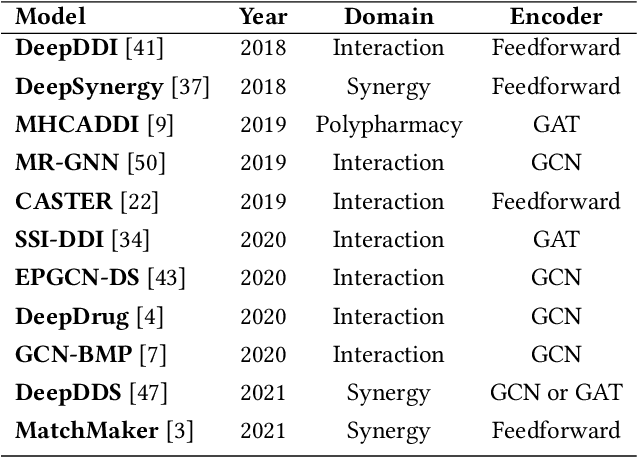
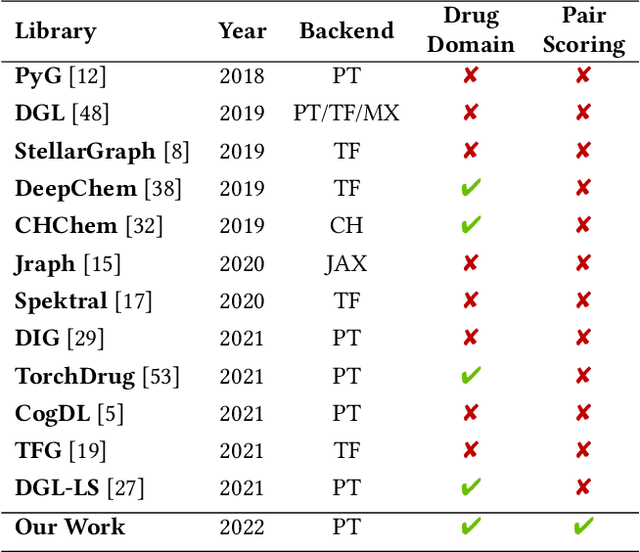
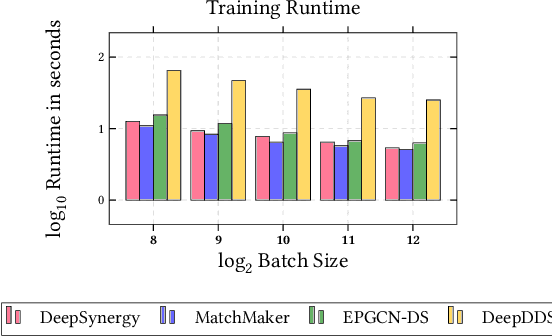
Abstract:In this paper, we introduce ChemicalX, a PyTorch-based deep learning library designed for providing a range of state of the art models to solve the drug pair scoring task. The primary objective of the library is to make deep drug pair scoring models accessible to machine learning researchers and practitioners in a streamlined framework.The design of ChemicalX reuses existing high level model training utilities, geometric deep learning, and deep chemistry layers from the PyTorch ecosystem. Our system provides neural network layers, custom pair scoring architectures, data loaders, and batch iterators for end users. We showcase these features with example code snippets and case studies to highlight the characteristics of ChemicalX. A range of experiments on real world drug-drug interaction, polypharmacy side effect, and combination synergy prediction tasks demonstrate that the models available in ChemicalX are effective at solving the pair scoring task. Finally, we show that ChemicalX could be used to train and score machine learning models on large drug pair datasets with hundreds of thousands of compounds on commodity hardware.
The Shapley Value in Machine Learning
Feb 11, 2022
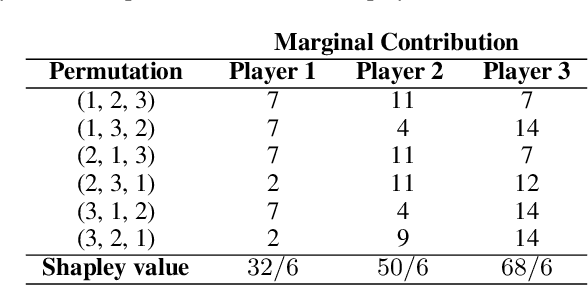
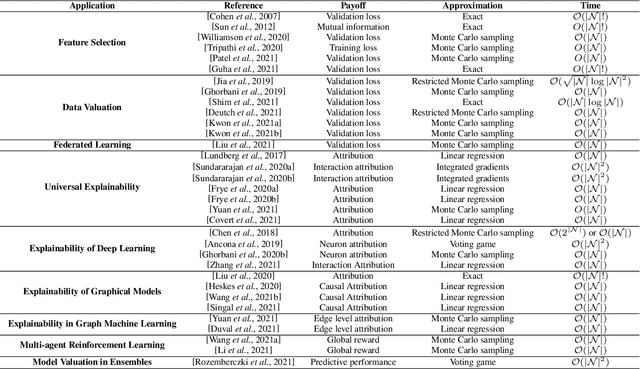
Abstract:Over the last few years, the Shapley value, a solution concept from cooperative game theory, has found numerous applications in machine learning. In this paper, we first discuss fundamental concepts of cooperative game theory and axiomatic properties of the Shapley value. Then we give an overview of the most important applications of the Shapley value in machine learning: feature selection, explainability, multi-agent reinforcement learning, ensemble pruning, and data valuation. We examine the most crucial limitations of the Shapley value and point out directions for future research.
Explainable Biomedical Recommendations via Reinforcement Learning Reasoning on Knowledge Graphs
Nov 20, 2021



Abstract:For Artificial Intelligence to have a greater impact in biology and medicine, it is crucial that recommendations are both accurate and transparent. In other domains, a neurosymbolic approach of multi-hop reasoning on knowledge graphs has been shown to produce transparent explanations. However, there is a lack of research applying it to complex biomedical datasets and problems. In this paper, the approach is explored for drug discovery to draw solid conclusions on its applicability. For the first time, we systematically apply it to multiple biomedical datasets and recommendation tasks with fair benchmark comparisons. The approach is found to outperform the best baselines by 21.7% on average whilst producing novel, biologically relevant explanations.
A Unified View of Relational Deep Learning for Drug Pair Scoring
Nov 14, 2021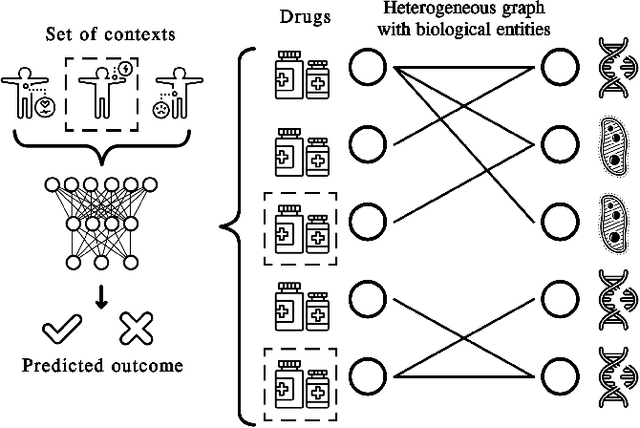
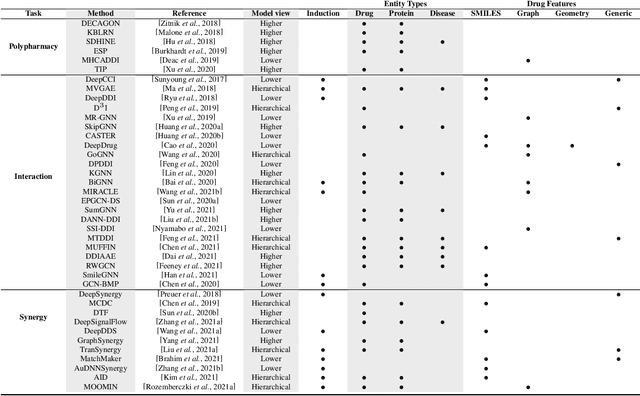
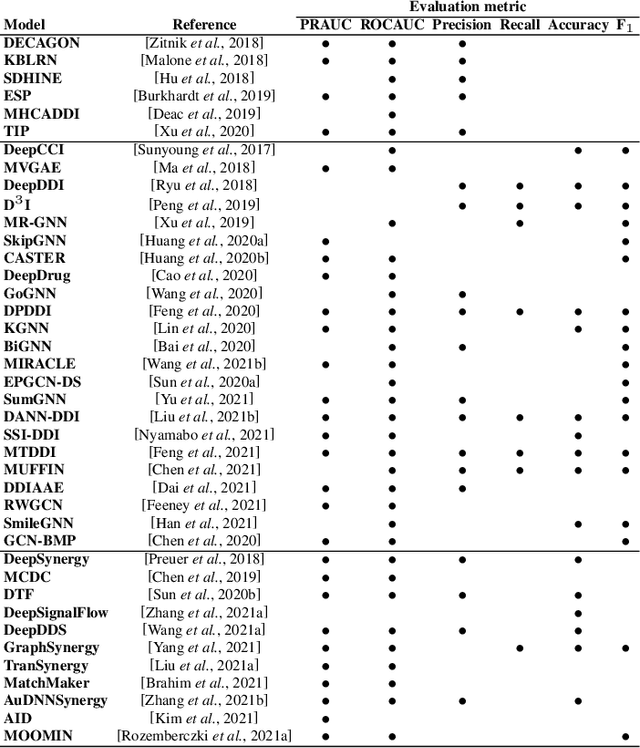
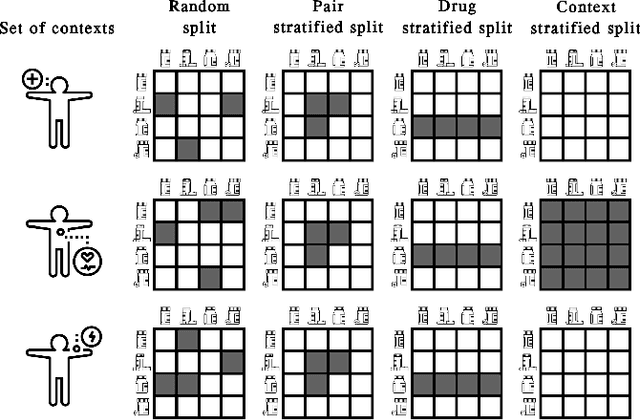
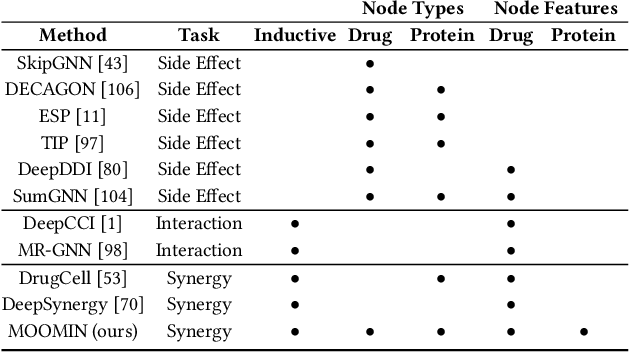
Abstract:In recent years, numerous machine learning models which attempt to solve polypharmacy side effect identification, drug-drug interaction prediction and combination therapy design tasks have been proposed. Here, we present a unified theoretical view of relational machine learning models which can address these tasks. We provide fundamental definitions, compare existing model architectures and discuss performance metrics, datasets and evaluation protocols. In addition, we emphasize possible high impact applications and important future research directions in this domain.
MOOMIN: Deep Molecular Omics Network for Anti-Cancer Drug Combination Therapy
Oct 28, 2021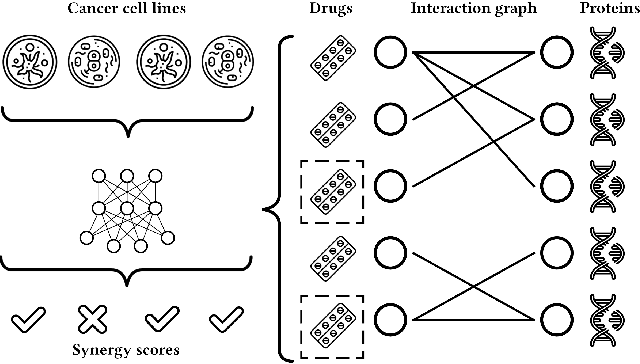
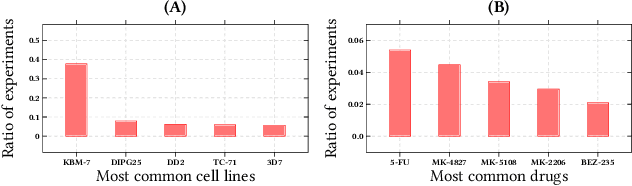

Abstract:We propose the molecular omics network (MOOMIN) a multimodal graph neural network that can predict the synergistic effect of drug combinations for cancer treatment. Our model captures the representation based on the context of drugs at multiple scales based on a drug-protein interaction network and metadata. Structural properties of the compounds and proteins are encoded to create vertex features for a message-passing scheme that operates on the bipartite interaction graph. Propagated messages form multi-resolution drug representations which we utilized to create drug pair descriptors. By conditioning the drug combination representations on the cancer cell type we define a synergy scoring function that can inductively score unseen pairs of drugs. Experimental results on the synergy scoring task demonstrate that MOOMIN outperforms state-of-the-art graph fingerprinting, proximity preserving node embedding, and existing deep learning approaches. Further results establish that the predictive performance of our model is robust to hyperparameter changes. We demonstrate that the model makes high-quality predictions over a wide range of cancer cell line tissues, out-of-sample predictions can be validated with external synergy databases, and that the proposed model is data-efficient at learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge