Gavin Edwards
Explainable Biomedical Recommendations via Reinforcement Learning Reasoning on Knowledge Graphs
Nov 20, 2021



Abstract:For Artificial Intelligence to have a greater impact in biology and medicine, it is crucial that recommendations are both accurate and transparent. In other domains, a neurosymbolic approach of multi-hop reasoning on knowledge graphs has been shown to produce transparent explanations. However, there is a lack of research applying it to complex biomedical datasets and problems. In this paper, the approach is explored for drug discovery to draw solid conclusions on its applicability. For the first time, we systematically apply it to multiple biomedical datasets and recommendation tasks with fair benchmark comparisons. The approach is found to outperform the best baselines by 21.7% on average whilst producing novel, biologically relevant explanations.
MOOMIN: Deep Molecular Omics Network for Anti-Cancer Drug Combination Therapy
Oct 28, 2021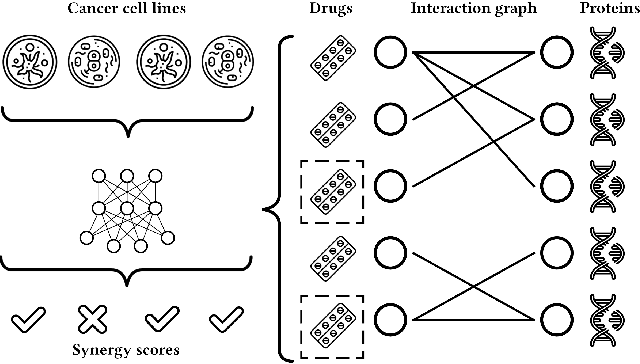

Abstract:We propose the molecular omics network (MOOMIN) a multimodal graph neural network that can predict the synergistic effect of drug combinations for cancer treatment. Our model captures the representation based on the context of drugs at multiple scales based on a drug-protein interaction network and metadata. Structural properties of the compounds and proteins are encoded to create vertex features for a message-passing scheme that operates on the bipartite interaction graph. Propagated messages form multi-resolution drug representations which we utilized to create drug pair descriptors. By conditioning the drug combination representations on the cancer cell type we define a synergy scoring function that can inductively score unseen pairs of drugs. Experimental results on the synergy scoring task demonstrate that MOOMIN outperforms state-of-the-art graph fingerprinting, proximity preserving node embedding, and existing deep learning approaches. Further results establish that the predictive performance of our model is robust to hyperparameter changes. We demonstrate that the model makes high-quality predictions over a wide range of cancer cell line tissues, out-of-sample predictions can be validated with external synergy databases, and that the proposed model is data-efficient at learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge