William L Hamilton
Understanding the Performance of Knowledge Graph Embeddings in Drug Discovery
Jun 07, 2021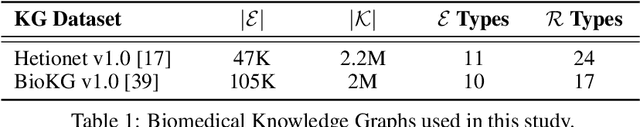
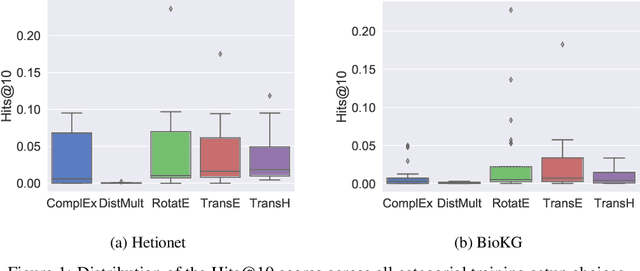
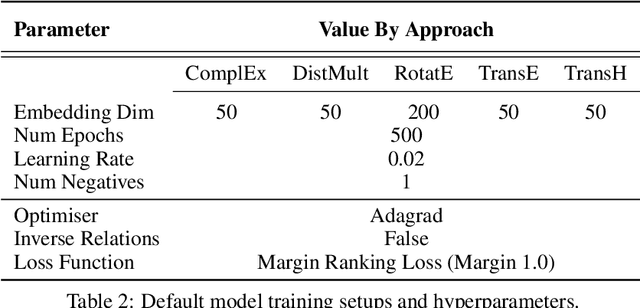
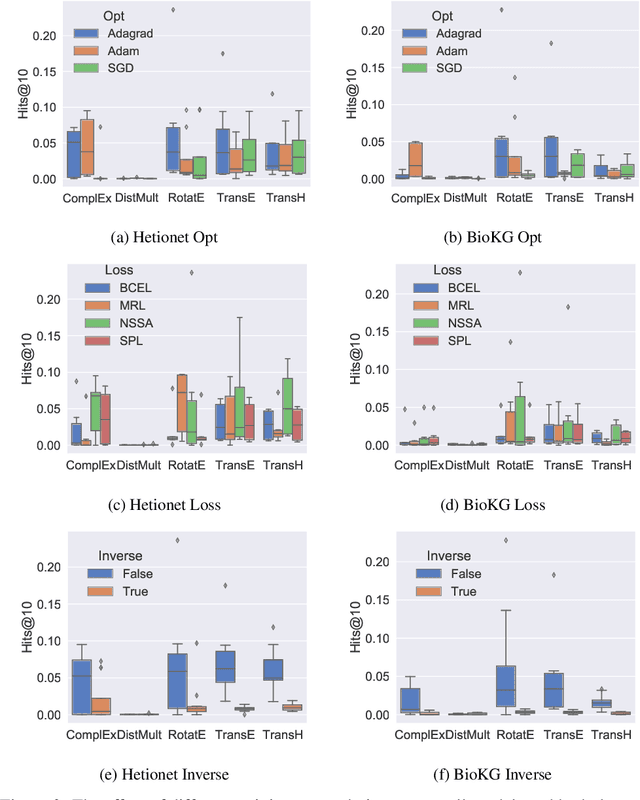
Abstract:Knowledge Graphs (KG) and associated Knowledge Graph Embedding (KGE) models have recently begun to be explored in the context of drug discovery and have the potential to assist in key challenges such as target identification. In the drug discovery domain, KGs can be employed as part of a process which can result in lab-based experiments being performed, or impact on other decisions, incurring significant time and financial costs and most importantly, ultimately influencing patient healthcare. For KGE models to have impact in this domain, a better understanding of not only of performance, but also the various factors which determine it, is required. In this study we investigate, over the course of many thousands of experiments, the predictive performance of five KGE models on two public drug discovery-oriented KGs. Our goal is not to focus on the best overall model or configuration, instead we take a deeper look at how performance can be affected by changes in the training setup, choice of hyperparameters, model parameter initialisation seed and different splits of the datasets. Our results highlight that these factors have significant impact on performance and can even affect the ranking of models. Indeed these factors should be reported along with model architectures to ensure complete reproducibility and fair comparisons of future work, and we argue this is critical for the acceptance of use, and impact of KGEs in a biomedical setting. To aid reproducibility of our own work, we release all experimentation code.
End-to-End Training of Neural Retrievers for Open-Domain Question Answering
Jan 02, 2021

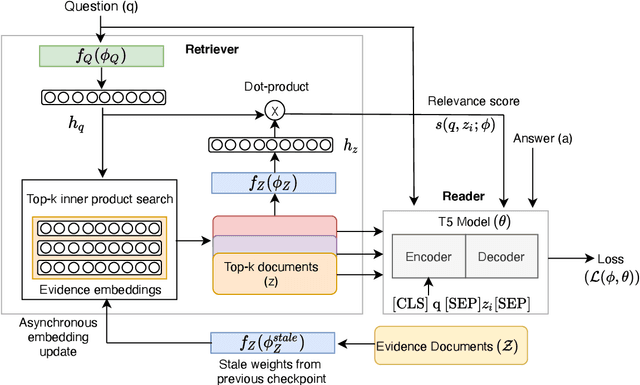
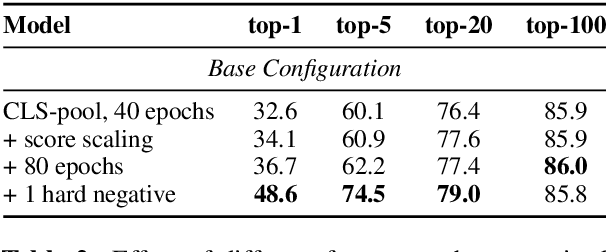
Abstract:Recent work on training neural retrievers for open-domain question answering (OpenQA) has employed both supervised and unsupervised approaches. However, it remains unclear how unsupervised and supervised methods can be used most effectively for neural retrievers. In this work, we systematically study retriever pre-training. We first propose an approach of unsupervised pre-training with the Inverse Cloze Task and masked salient spans, followed by supervised finetuning using question-context pairs. This approach leads to absolute gains of 2+ points over the previous best result in the top-20 retrieval accuracy on Natural Questions and TriviaQA datasets. We also explore two approaches for end-to-end supervised training of the reader and retriever components in OpenQA models. In the first approach, the reader considers each retrieved document separately while in the second approach, the reader considers all the retrieved documents together. Our experiments demonstrate the effectiveness of these approaches as we obtain new state-of-the-art results. On the Natural Questions dataset, we obtain a top-20 retrieval accuracy of 84, an improvement of 5 points over the recent DPR model. In addition, we achieve good results on answer extraction, outperforming recent models like REALM and RAG by 3+ points. We further scale up end-to-end training to large models and show consistent gains in performance over smaller models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge