Rowan Swiers
Batched Online Contextual Sparse Bandits with Sequential Inclusion of Features
Sep 13, 2024Abstract:Multi-armed Bandits (MABs) are increasingly employed in online platforms and e-commerce to optimize decision making for personalized user experiences. In this work, we focus on the Contextual Bandit problem with linear rewards, under conditions of sparsity and batched data. We address the challenge of fairness by excluding irrelevant features from decision-making processes using a novel algorithm, Online Batched Sequential Inclusion (OBSI), which sequentially includes features as confidence in their impact on the reward increases. Our experiments on synthetic data show the superior performance of OBSI compared to other algorithms in terms of regret, relevance of features used, and compute.
Understanding the Performance of Knowledge Graph Embeddings in Drug Discovery
Jun 07, 2021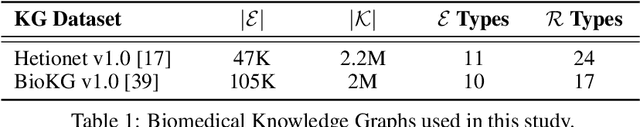
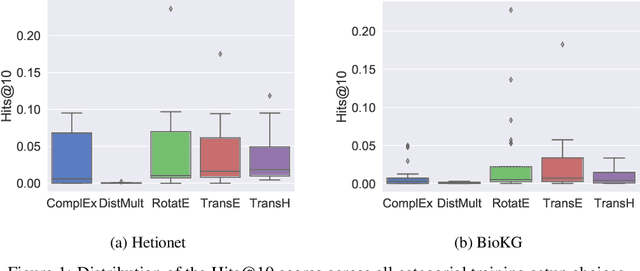
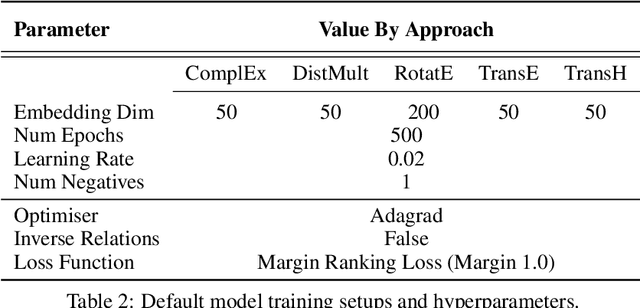
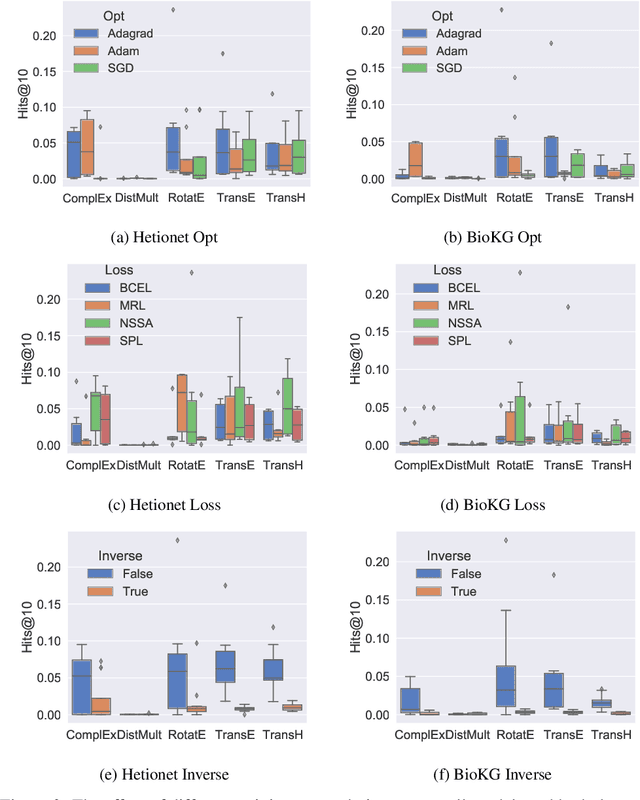
Abstract:Knowledge Graphs (KG) and associated Knowledge Graph Embedding (KGE) models have recently begun to be explored in the context of drug discovery and have the potential to assist in key challenges such as target identification. In the drug discovery domain, KGs can be employed as part of a process which can result in lab-based experiments being performed, or impact on other decisions, incurring significant time and financial costs and most importantly, ultimately influencing patient healthcare. For KGE models to have impact in this domain, a better understanding of not only of performance, but also the various factors which determine it, is required. In this study we investigate, over the course of many thousands of experiments, the predictive performance of five KGE models on two public drug discovery-oriented KGs. Our goal is not to focus on the best overall model or configuration, instead we take a deeper look at how performance can be affected by changes in the training setup, choice of hyperparameters, model parameter initialisation seed and different splits of the datasets. Our results highlight that these factors have significant impact on performance and can even affect the ranking of models. Indeed these factors should be reported along with model architectures to ensure complete reproducibility and fair comparisons of future work, and we argue this is critical for the acceptance of use, and impact of KGEs in a biomedical setting. To aid reproducibility of our own work, we release all experimentation code.
Predicting Potential Drug Targets Using Tensor Factorisation and Knowledge Graph Embeddings
May 20, 2021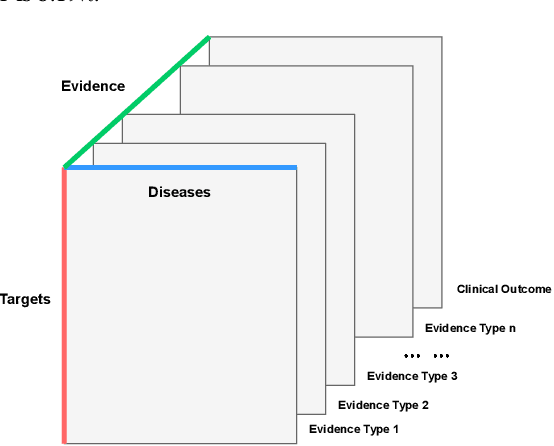
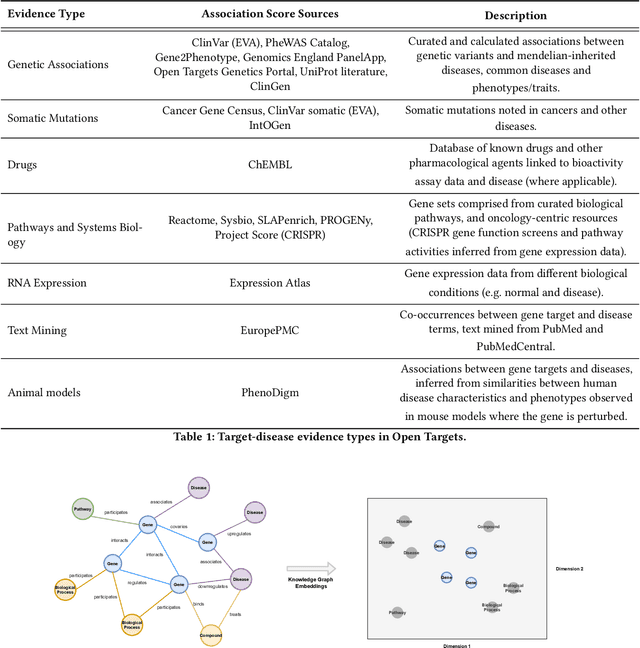
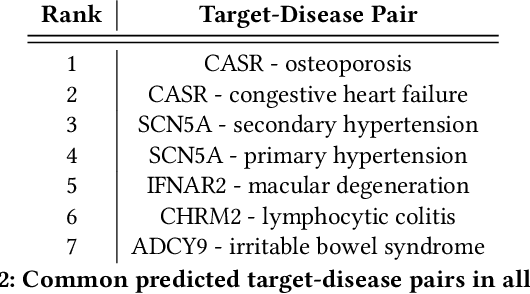
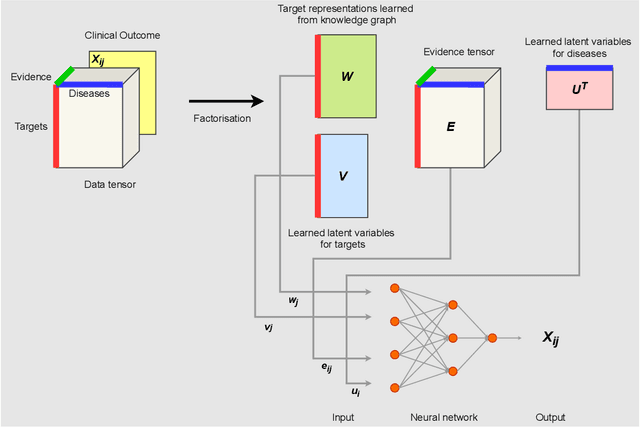
Abstract:The drug discovery and development process is a long and expensive one, costing over 1 billion USD on average per drug and taking 10-15 years. To reduce the high levels of attrition throughout the process, there has been a growing interest in applying machine learning methodologies to various stages of drug discovery process in the recent decade, including at the earliest stage - identification of druggable disease genes. In this paper, we have developed a new tensor factorisation model to predict potential drug targets (i.e.,genes or proteins) for diseases. We created a three dimensional tensor which consists of 1,048 targets, 860 diseases and 230,011 evidence attributes and clinical outcomes connecting them, using data extracted from the Open Targets and PharmaProjects databases. We enriched the data with gene representations learned from a drug discovery-oriented knowledge graph and applied our proposed method to predict the clinical outcomes for unseen target and dis-ease pairs. We designed three evaluation strategies to measure the prediction performance and benchmarked several commonly used machine learning classifiers together with matrix and tensor factorisation methods. The result shows that incorporating knowledge graph embeddings significantly improves the prediction accuracy and that training tensor factorisation alongside a dense neural network outperforms other methods. In summary, our framework combines two actively studied machine learning approaches to disease target identification, tensor factorisation and knowledge graph representation learning, which could be a promising avenue for further exploration in data-driven drug discovery.
A Review of Biomedical Datasets Relating to Drug Discovery: A Knowledge Graph Perspective
Feb 26, 2021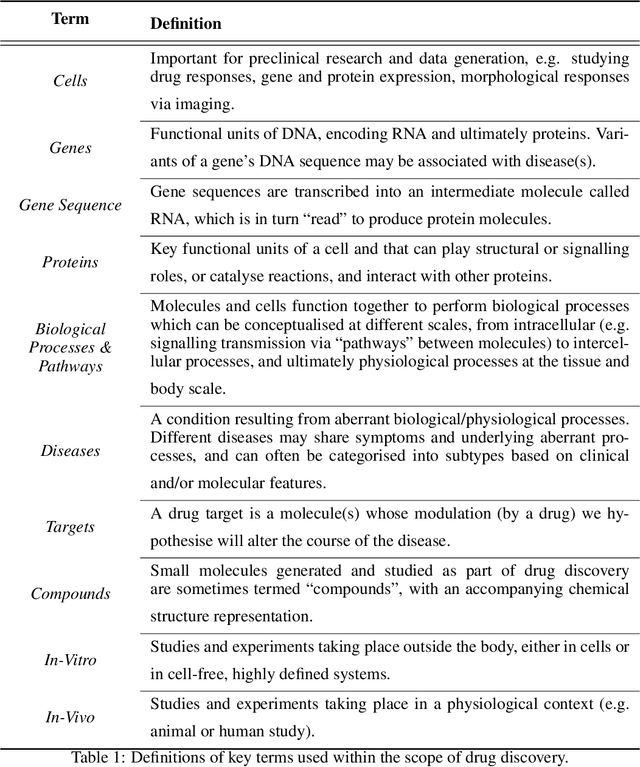
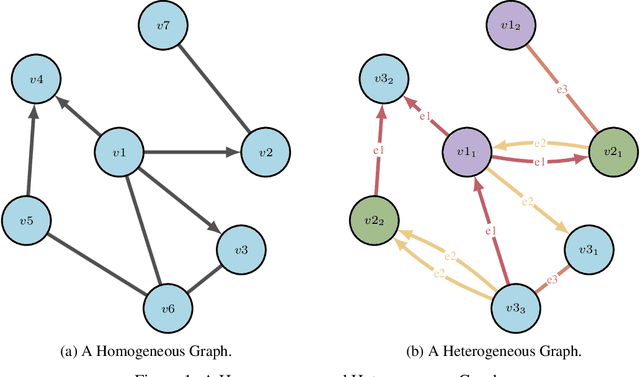
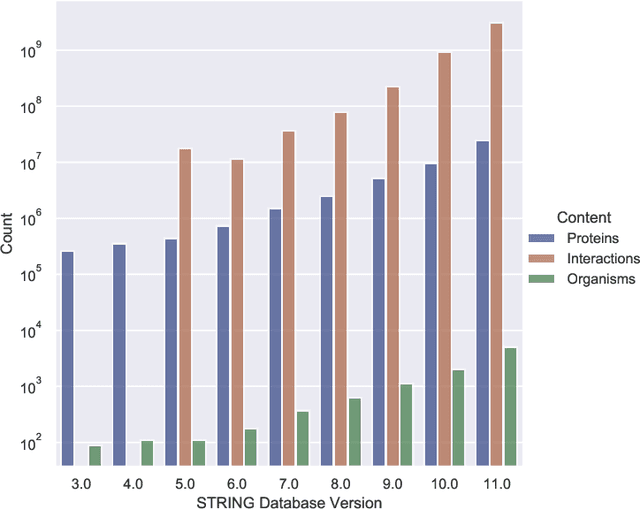
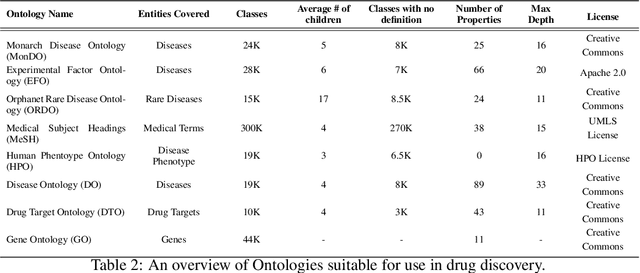
Abstract:Drug discovery and development is an extremely complex process, with high attrition contributing to the costs of delivering new medicines to patients. Recently, various machine learning approaches have been proposed and investigated to help improve the effectiveness and speed of multiple stages of the drug discovery pipeline. Among these techniques, it is especially those using Knowledge Graphs that are proving to have considerable promise across a range of tasks, including drug repurposing, drug toxicity prediction and target gene-disease prioritisation. In such a knowledge graph-based representation of drug discovery domains, crucial elements including genes, diseases and drugs are represented as entities or vertices, whilst relationships or edges between them indicate some level of interaction. For example, an edge between a disease and drug entity might represent a successful clinical trial, or an edge between two drug entities could indicate a potentially harmful interaction. In order to construct high-quality and ultimately informative knowledge graphs however, suitable data and information is of course required. In this review, we detail publicly available primary data sources containing information suitable for use in constructing various drug discovery focused knowledge graphs. We aim to help guide machine learning and knowledge graph practitioners who are interested in applying new techniques to the drug discovery field, but who may be unfamiliar with the relevant data sources. Overall we hope this review will help motivate more machine learning researchers to explore combining knowledge graphs and machine learning to help solve key and emerging questions in the drug discovery domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge