Kaushal Paneri
Leveraging Structured Biological Knowledge for Counterfactual Inference: a Case Study of Viral Pathogenesis
Jan 13, 2021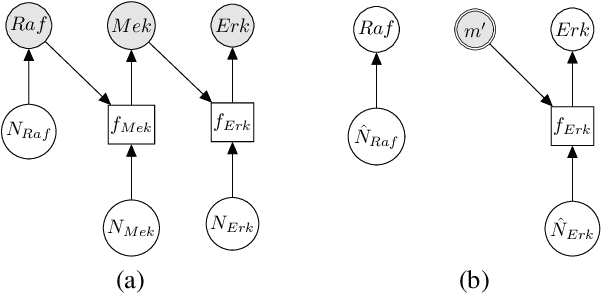
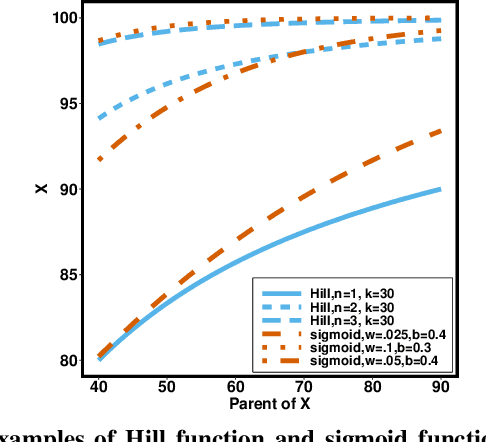
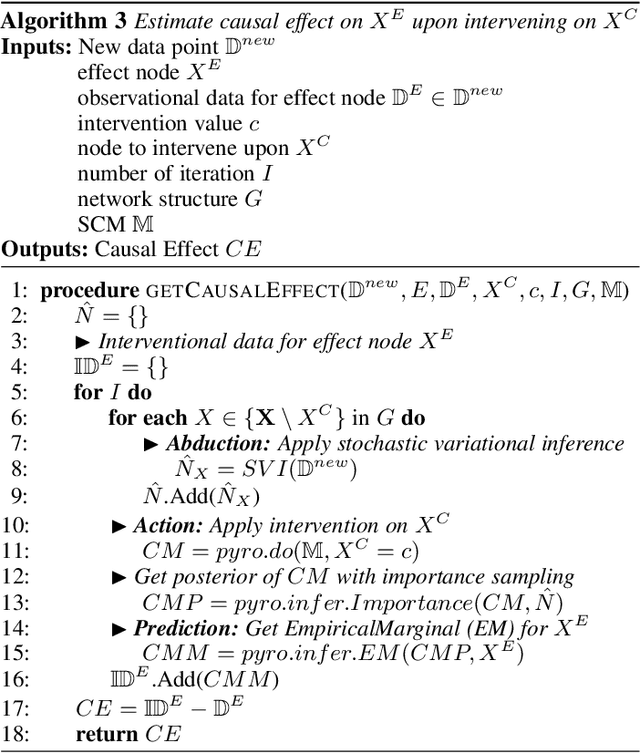
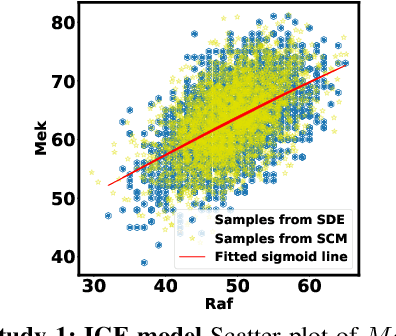
Abstract:Counterfactual inference is a useful tool for comparing outcomes of interventions on complex systems. It requires us to represent the system in form of a structural causal model, complete with a causal diagram, probabilistic assumptions on exogenous variables, and functional assignments. Specifying such models can be extremely difficult in practice. The process requires substantial domain expertise, and does not scale easily to large systems, multiple systems, or novel system modifications. At the same time, many application domains, such as molecular biology, are rich in structured causal knowledge that is qualitative in nature. This manuscript proposes a general approach for querying a causal biological knowledge graph, and converting the qualitative result into a quantitative structural causal model that can learn from data to answer the question. We demonstrate the feasibility, accuracy and versatility of this approach using two case studies in systems biology. The first demonstrates the appropriateness of the underlying assumptions and the accuracy of the results. The second demonstrates the versatility of the approach by querying a knowledge base for the molecular determinants of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced cytokine storm, and performing counterfactual inference to estimate the causal effect of medical countermeasures for severely ill patients.
Integrating Markov processes with structural causal modeling enables counterfactual inference in complex systems
Nov 06, 2019


Abstract:This manuscript contributes a general and practical framework for casting a Markov process model of a system at equilibrium as a structural causal model, and carrying out counterfactual inference. Markov processes mathematically describe the mechanisms in the system, and predict the system's equilibrium behavior upon intervention, but do not support counterfactual inference. In contrast, structural causal models support counterfactual inference, but do not identify the mechanisms. This manuscript leverages the benefits of both approaches. We define the structural causal models in terms of the parameters and the equilibrium dynamics of the Markov process models, and counterfactual inference flows from these settings. The proposed approach alleviates the identifiability drawback of the structural causal models, in that the counterfactual inference is consistent with the counterfactual trajectories simulated from the Markov process model. We showcase the benefits of this framework in case studies of complex biomolecular systems with nonlinear dynamics. We illustrate that, in presence of Markov process model misspecification, counterfactual inference leverages prior data, and therefore estimates the outcome of an intervention more accurately than a direct simulation.
Crop Planning using Stochastic Visual Optimization
Oct 25, 2017

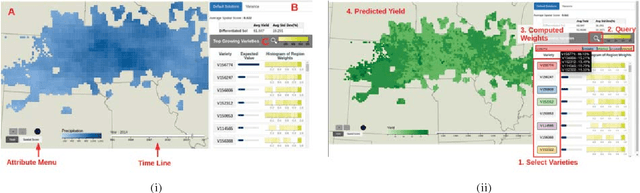

Abstract:As the world population increases and arable land decreases, it becomes vital to improve the productivity of the agricultural land available. Given the weather and soil properties, farmers need to take critical decisions such as which seed variety to plant and in what proportion, in order to maximize productivity. These decisions are irreversible and any unusual behavior of external factors, such as weather, can have catastrophic impact on the productivity of crop. A variety which is highly desirable to a farmer might be unavailable or in short supply, therefore, it is very critical to evaluate which variety or varieties are more likely to be chosen by farmers from a growing region in order to meet demand. In this paper, we present our visual analytics tool, ViSeed, showcased on the data given in Syngenta 2016 crop data challenge 1 . This tool helps to predict optimal soybean seed variety or mix of varieties in appropriate proportions which is more likely to be chosen by farmers from a growing region. It also allows to analyse solutions generated from our approach and helps in the decision making process by providing insightful visualizations
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge