Myocardial
Papers and Code
Point Tracking as a Temporal Cue for Robust Myocardial Segmentation in Echocardiography Videos
Jan 14, 2026Purpose: Myocardium segmentation in echocardiography videos is a challenging task due to low contrast, noise, and anatomical variability. Traditional deep learning models either process frames independently, ignoring temporal information, or rely on memory-based feature propagation, which accumulates error over time. Methods: We propose Point-Seg, a transformer-based segmentation framework that integrates point tracking as a temporal cue to ensure stable and consistent segmentation of myocardium across frames. Our method leverages a point-tracking module trained on a synthetic echocardiography dataset to track key anatomical landmarks across video sequences. These tracked trajectories provide an explicit motion-aware signal that guides segmentation, reducing drift and eliminating the need for memory-based feature accumulation. Additionally, we incorporate a temporal smoothing loss to further enhance temporal consistency across frames. Results: We evaluate our approach on both public and private echocardiography datasets. Experimental results demonstrate that Point-Seg has statistically similar accuracy in terms of Dice to state-of-the-art segmentation models in high quality echo data, while it achieves better segmentation accuracy in lower quality echo with improved temporal stability. Furthermore, Point-Seg has the key advantage of pixel-level myocardium motion information as opposed to other segmentation methods. Such information is essential in the computation of other downstream tasks such as myocardial strain measurement and regional wall motion abnormality detection. Conclusion: Point-Seg demonstrates that point tracking can serve as an effective temporal cue for consistent video segmentation, offering a reliable and generalizable approach for myocardium segmentation in echocardiography videos. The code is available at https://github.com/DeepRCL/PointSeg.
Contextual Discrepancy-Aware Contrastive Learning for Robust Medical Time Series Diagnosis in Small-Sample Scenarios
Jan 12, 2026Medical time series data, such as EEG and ECG, are vital for diagnosing neurological and cardiovascular diseases. However, their precise interpretation faces significant challenges due to high annotation costs, leading to data scarcity, and the limitations of traditional contrastive learning in capturing complex temporal patterns. To address these issues, we propose CoDAC (Contextual Discrepancy-Aware Contrastive learning), a novel framework that enhances diagnostic accuracy and generalization, particularly in small-sample settings. CoDAC leverages external healthy data and introduces a Contextual Discrepancy Estimator (CDE), built upon a Transformer-based Autoencoder, to precisely quantify abnormal signals through context-aware anomaly scores. These scores dynamically inform a Dynamic Multi-views Contrastive Framework (DMCF), which adaptively weights different temporal views to focus contrastive learning on diagnostically relevant, discrepant regions. Our encoder combines dilated convolutions with multi-head attention for robust feature extraction. Comprehensive experiments on Alzheimer's Disease EEG, Parkinson's Disease EEG, and Myocardial Infarction ECG datasets demonstrate CoDAC's superior performance across all metrics, consistently outperforming state-of-the-art baselines, especially under low label availability. Ablation studies further validate the critical contributions of CDE and DMCF. CoDAC offers a robust and interpretable solution for medical time series diagnosis, effectively mitigating data scarcity challenges.
Transformer-Based Multi-Modal Temporal Embeddings for Explainable Metabolic Phenotyping in Type 1 Diabetes
Jan 07, 2026Type 1 diabetes (T1D) is a highly metabolically heterogeneous disease that cannot be adequately characterized by conventional biomarkers such as glycated hemoglobin (HbA1c). This study proposes an explainable deep learning framework that integrates continuous glucose monitoring (CGM) data with laboratory profiles to learn multimodal temporal embeddings of individual metabolic status. Temporal dependencies across modalities are modeled using a transformer encoder, while latent metabolic phenotypes are identified via Gaussian mixture modeling. Model interpretability is achieved through transformer attention visualization and SHAP-based feature attribution. Five latent metabolic phenotypes, ranging from metabolic stability to elevated cardiometabolic risk, were identified among 577 individuals with T1D. These phenotypes exhibit distinct biochemical profiles, including differences in glycemic control, lipid metabolism, renal markers, and thyrotropin (TSH) levels. Attention analysis highlights glucose variability as a dominant temporal factor, while SHAP analysis identifies HbA1c, triglycerides, cholesterol, creatinine, and TSH as key contributors to phenotype differentiation. Phenotype membership shows statistically significant, albeit modest, associations with hypertension, myocardial infarction, and heart failure. Overall, this explainable multimodal temporal embedding framework reveals physiologically coherent metabolic subgroups in T1D and supports risk stratification beyond single biomarkers.
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Deep learning-enhanced dual-mode multiplexed optical sensor for point-of-care diagnostics of cardiovascular diseases
Dec 24, 2025Rapid and accessible cardiac biomarker testing is essential for the timely diagnosis and risk assessment of myocardial infarction (MI) and heart failure (HF), two interrelated conditions that frequently coexist and drive recurrent hospitalizations with high mortality. However, current laboratory and point-of-care testing systems are limited by long turnaround times, narrow dynamic ranges for the tested biomarkers, and single-analyte formats that fail to capture the complexity of cardiovascular disease. Here, we present a deep learning-enhanced dual-mode multiplexed vertical flow assay (xVFA) with a portable optical reader and a neural network-based quantification pipeline. This optical sensor integrates colorimetric and chemiluminescent detection within a single paper-based cartridge to complementarily cover a large dynamic range (spanning ~6 orders of magnitude) for both low- and high-abundance biomarkers, while maintaining quantitative accuracy. Using 50 uL of serum, the optical sensor simultaneously quantifies cardiac troponin I (cTnI), creatine kinase-MB (CK-MB), and N-terminal pro-B-type natriuretic peptide (NT-proBNP) within 23 min. The xVFA achieves sub-pg/mL sensitivity for cTnI and sub-ng/mL sensitivity for CK-MB and NT-proBNP, spanning the clinically relevant ranges for these biomarkers. Neural network models trained and blindly tested on 92 patient serum samples yielded a robust quantification performance (Pearson's r > 0.96 vs. reference assays). By combining high sensitivity, multiplexing, and automation in a compact and cost-effective optical sensor format, the dual-mode xVFA enables rapid and quantitative cardiovascular diagnostics at the point of care.
SCAR: Semantic Cardiac Adversarial Representation via Spatiotemporal Manifold Optimization in ECG
Dec 19, 2025



Deep learning models for Electrocardiogram (ECG) analysis have achieved expert-level performance but remain vulnerable to adversarial attacks. However, applying Universal Adversarial Perturbations (UAP) to ECG signals presents a unique challenge: standard imperceptible noise constraints (e.g., 10 uV) fail to generate effective universal attacks due to the high inter-subject variability of cardiac waveforms. Furthermore, traditional "invisible" attacks are easily dismissed by clinicians as technical artifacts, failing to compromise the human-in-the-loop diagnostic pipeline. In this study, we propose SCAR (Semantic Cardiac Adversarial Representation), a novel UAP framework tailored to bypass the clinical "Human Firewall." Unlike traditional approaches, SCAR integrates spatiotemporal smoothing (W=25, approx. 50ms), spectral consistency (<15 Hz), and anatomical amplitude constraints (<0.2 mV) directly into the gradient optimization manifold. Results: We benchmarked SCAR against a rigorous baseline (Standard Universal DeepFool with post-hoc physiological filtering). While the baseline suffers a performance collapse (~16% success rate on transfer tasks), SCAR maintains robust transferability (58.09% on ResNet) and achieves 82.46% success on the source model. Crucially, clinical analysis reveals an emergent targeted behavior: SCAR specifically converges to forging Myocardial Infarction features (90.2% misdiagnosis) by mathematically reconstructing pathological ST-segment elevations. Finally, we demonstrate that SCAR serves a dual purpose: it not only functions as a robust data augmentation strategy for Hybrid Adversarial Training, offering optimal clinical defense, but also provides effective educational samples for training clinicians to recognize low-cost, AI-targeted semantic forgeries.
Error Bound Analysis of Physics-Informed Neural Networks-Driven T2 Quantification in Cardiac Magnetic Resonance Imaging
Dec 16, 2025



Physics-Informed Neural Networks (PINN) are emerging as a promising approach for quantitative parameter estimation of Magnetic Resonance Imaging (MRI). While existing deep learning methods can provide an accurate quantitative estimation of the T2 parameter, they still require large amounts of training data and lack theoretical support and a recognized gold standard. Thus, given the absence of PINN-based approaches for T2 estimation, we propose embedding the fundamental physics of MRI, the Bloch equation, in the loss of PINN, which is solely based on target scan data and does not require a pre-defined training database. Furthermore, by deriving rigorous upper bounds for both the T2 estimation error and the generalization error of the Bloch equation solution, we establish a theoretical foundation for evaluating the PINN's quantitative accuracy. Even without access to the ground truth or a gold standard, this theory enables us to estimate the error with respect to the real quantitative parameter T2. The accuracy of T2 mapping and the validity of the theoretical analysis are demonstrated on a numerical cardiac model and a water phantom, where our method exhibits excellent quantitative precision in the myocardial T2 range. Clinical applicability is confirmed in 94 acute myocardial infarction (AMI) patients, achieving low-error quantitative T2 estimation under the theoretical error bound, highlighting the robustness and potential of PINN.
Anatomy Guided Coronary Artery Segmentation from CCTA Using Spatial Frequency Joint Modeling
Dec 14, 2025Accurate coronary artery segmentation from coronary computed tomography angiography is essential for quantitative coronary analysis and clinical decision support. Nevertheless, reliable segmentation remains challenging because of small vessel calibers, complex branching, blurred boundaries, and myocardial interference. We propose a coronary artery segmentation framework that integrates myocardial anatomical priors, structure aware feature encoding, and three dimensional wavelet inverse wavelet transformations. Myocardial priors and residual attention based feature enhancement are incorporated during encoding to strengthen coronary structure representation. Wavelet inverse wavelet based downsampling and upsampling enable joint spatial frequency modeling and preserve multi scale structural consistency, while a multi scale feature fusion module integrates semantic and geometric information in the decoding stage. The model is trained and evaluated on the public ImageCAS dataset using a 3D overlapping patch based strategy with a 7:1:2 split for training, validation, and testing. Experimental results demonstrate that the proposed method achieves a Dice coefficient of 0.8082, Sensitivity of 0.7946, Precision of 0.8471, and an HD95 of 9.77 mm, outperforming several mainstream segmentation models. Ablation studies further confirm the complementary contributions of individual components. The proposed method enables more stable and consistent coronary artery segmentation under complex geometric conditions, providing reliable segmentation results for subsequent coronary structure analysis tasks.
Knowledge-Informed Automatic Feature Extraction via Collaborative Large Language Model Agents
Nov 19, 2025The performance of machine learning models on tabular data is critically dependent on high-quality feature engineering. While Large Language Models (LLMs) have shown promise in automating feature extraction (AutoFE), existing methods are often limited by monolithic LLM architectures, simplistic quantitative feedback, and a failure to systematically integrate external domain knowledge. This paper introduces Rogue One, a novel, LLM-based multi-agent framework for knowledge-informed automatic feature extraction. Rogue One operationalizes a decentralized system of three specialized agents-Scientist, Extractor, and Tester-that collaborate iteratively to discover, generate, and validate predictive features. Crucially, the framework moves beyond primitive accuracy scores by introducing a rich, qualitative feedback mechanism and a "flooding-pruning" strategy, allowing it to dynamically balance feature exploration and exploitation. By actively incorporating external knowledge via an integrated retrieval-augmented (RAG) system, Rogue One generates features that are not only statistically powerful but also semantically meaningful and interpretable. We demonstrate that Rogue One significantly outperforms state-of-the-art methods on a comprehensive suite of 19 classification and 9 regression datasets. Furthermore, we show qualitatively that the system surfaces novel, testable hypotheses, such as identifying a new potential biomarker in the myocardial dataset, underscoring its utility as a tool for scientific discovery.
Seeing Beyond the Image: ECG and Anatomical Knowledge-Guided Myocardial Scar Segmentation from Late Gadolinium-Enhanced Images
Nov 18, 2025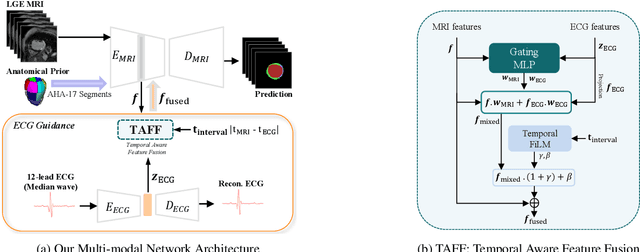
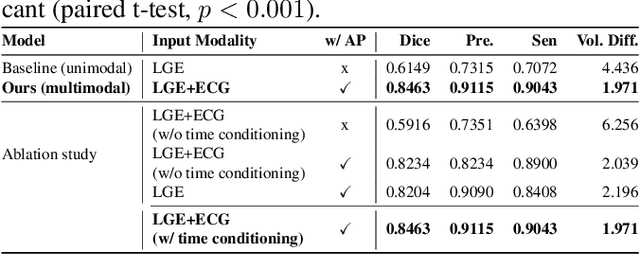
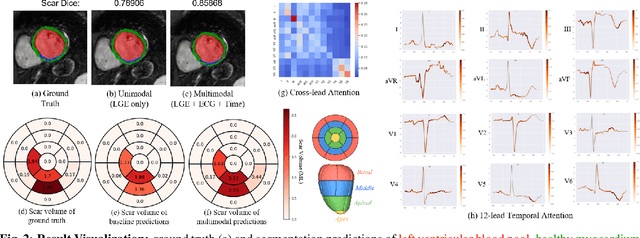
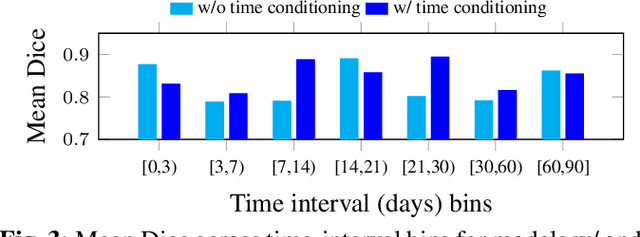
Accurate segmentation of myocardial scar from late gadolinium enhanced (LGE) cardiac MRI is essential for evaluating tissue viability, yet remains challenging due to variable contrast and imaging artifacts. Electrocardiogram (ECG) signals provide complementary physiological information, as conduction abnormalities can help localize or suggest scarred myocardial regions. In this work, we propose a novel multimodal framework that integrates ECG-derived electrophysiological information with anatomical priors from the AHA-17 atlas for physiologically consistent LGE-based scar segmentation. As ECGs and LGE-MRIs are not acquired simultaneously, we introduce a Temporal Aware Feature Fusion (TAFF) mechanism that dynamically weights and fuses features based on their acquisition time difference. Our method was evaluated on a clinical dataset and achieved substantial gains over the state-of-the-art image-only baseline (nnU-Net), increasing the average Dice score for scars from 0.6149 to 0.8463 and achieving high performance in both precision (0.9115) and sensitivity (0.9043). These results show that integrating physiological and anatomical knowledge allows the model to "see beyond the image", setting a new direction for robust and physiologically grounded cardiac scar segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge