Caizhi Liao
BEAT-Net: Injecting Biomimetic Spatio-Temporal Priors for Interpretable ECG Classification
Jan 12, 2026Abstract:Although deep learning has advanced automated electrocardiogram (ECG) diagnosis, prevalent supervised methods typically treat recordings as undifferentiated one-dimensional (1D) signals or two-dimensional (2D) images. This formulation compels models to learn physiological structures implicitly, resulting in data inefficiency and opacity that diverge from medical reasoning. To address these limitations, we propose BEAT-Net, a Biomimetic ECG Analysis with Tokenization framework that reformulates the problem as a language modeling task. Utilizing a QRS tokenization strategy to transform continuous signals into biologically aligned heartbeat sequences, the architecture explicitly decomposes cardiac physiology through specialized encoders that extract local beat morphology while normalizing spatial lead perspectives and modeling temporal rhythm dependencies. Evaluations across three large-scale benchmarks demonstrate that BEAT-Net matches the diagnostic accuracy of dominant convolutional neural network (CNN) architectures while substantially improving robustness. The framework exhibits exceptional data efficiency, recovering fully supervised performance using only 30 to 35 percent of annotated data. Moreover, learned attention mechanisms provide inherent interpretability by spontaneously reproducing clinical heuristics, such as Lead II prioritization for rhythm analysis, without explicit supervision. These findings indicate that integrating biological priors offers a computationally efficient and interpretable alternative to data-intensive large-scale pre-training.
CPR: Causal Physiological Representation Learning for Robust ECG Analysis under Distribution Shifts
Dec 31, 2025Abstract:Deep learning models for Electrocardiogram (ECG) diagnosis have achieved remarkable accuracy but exhibit fragility against adversarial perturbations, particularly Smooth Adversarial Perturbations (SAP) that mimic biological morphology. Existing defenses face a critical dilemma: Adversarial Training (AT) provides robustness but incurs a prohibitive computational burden, while certified methods like Randomized Smoothing (RS) introduce significant inference latency, rendering them impractical for real-time clinical monitoring. We posit that this vulnerability stems from the models' reliance on non-robust spurious correlations rather than invariant pathological features. To address this, we propose Causal Physiological Representation Learning (CPR). Unlike standard denoising approaches that operate without semantic constraints, CPR incorporates a Physiological Structural Prior within a causal disentanglement framework. By modeling ECG generation via a Structural Causal Model (SCM), CPR enforces a structural intervention that strictly separates invariant pathological morphology (P-QRS-T complex) from non-causal artifacts. Empirical results on PTB-XL demonstrate that CPR significantly outperforms standard clinical preprocessing methods. Specifically, under SAP attacks, CPR achieves an F1 score of 0.632, surpassing Median Smoothing (0.541 F1) by 9.1%. Crucially, CPR matches the certified robustness of Randomized Smoothing while maintaining single-pass inference efficiency, offering a superior trade-off between robustness, efficiency, and clinical interpretability.
SCAR: Semantic Cardiac Adversarial Representation via Spatiotemporal Manifold Optimization in ECG
Dec 19, 2025



Abstract:Deep learning models for Electrocardiogram (ECG) analysis have achieved expert-level performance but remain vulnerable to adversarial attacks. However, applying Universal Adversarial Perturbations (UAP) to ECG signals presents a unique challenge: standard imperceptible noise constraints (e.g., 10 uV) fail to generate effective universal attacks due to the high inter-subject variability of cardiac waveforms. Furthermore, traditional "invisible" attacks are easily dismissed by clinicians as technical artifacts, failing to compromise the human-in-the-loop diagnostic pipeline. In this study, we propose SCAR (Semantic Cardiac Adversarial Representation), a novel UAP framework tailored to bypass the clinical "Human Firewall." Unlike traditional approaches, SCAR integrates spatiotemporal smoothing (W=25, approx. 50ms), spectral consistency (<15 Hz), and anatomical amplitude constraints (<0.2 mV) directly into the gradient optimization manifold. Results: We benchmarked SCAR against a rigorous baseline (Standard Universal DeepFool with post-hoc physiological filtering). While the baseline suffers a performance collapse (~16% success rate on transfer tasks), SCAR maintains robust transferability (58.09% on ResNet) and achieves 82.46% success on the source model. Crucially, clinical analysis reveals an emergent targeted behavior: SCAR specifically converges to forging Myocardial Infarction features (90.2% misdiagnosis) by mathematically reconstructing pathological ST-segment elevations. Finally, we demonstrate that SCAR serves a dual purpose: it not only functions as a robust data augmentation strategy for Hybrid Adversarial Training, offering optimal clinical defense, but also provides effective educational samples for training clinicians to recognize low-cost, AI-targeted semantic forgeries.
A deep learning-enabled smart garment for versatile sleep behaviour monitoring
Aug 01, 2024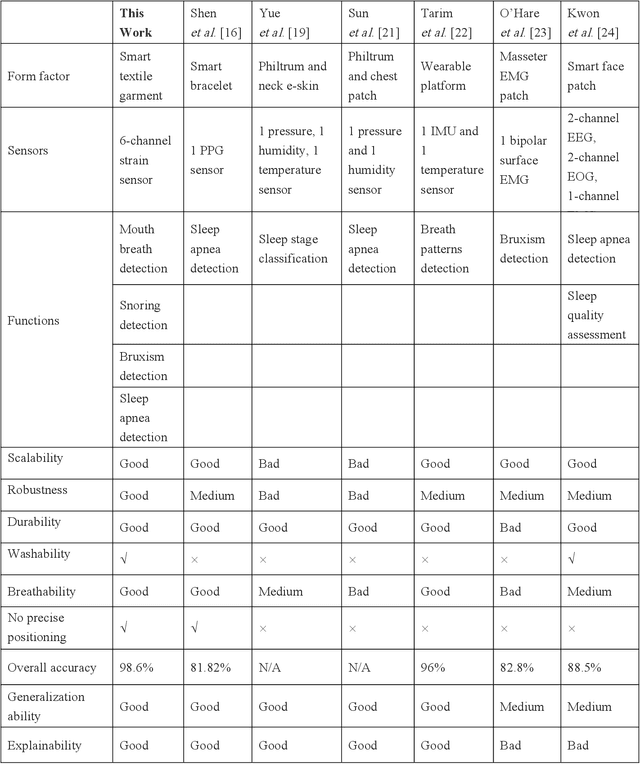
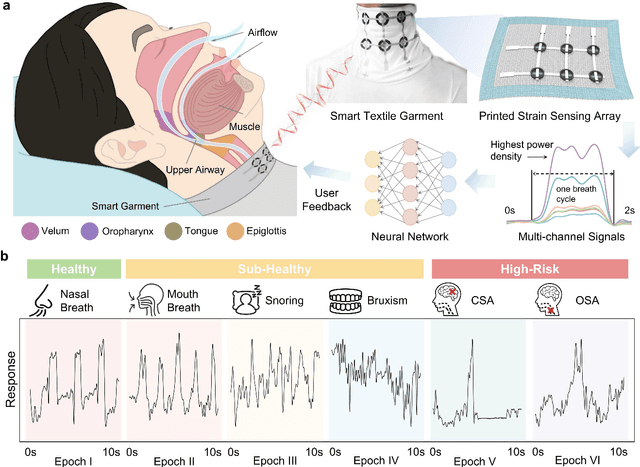
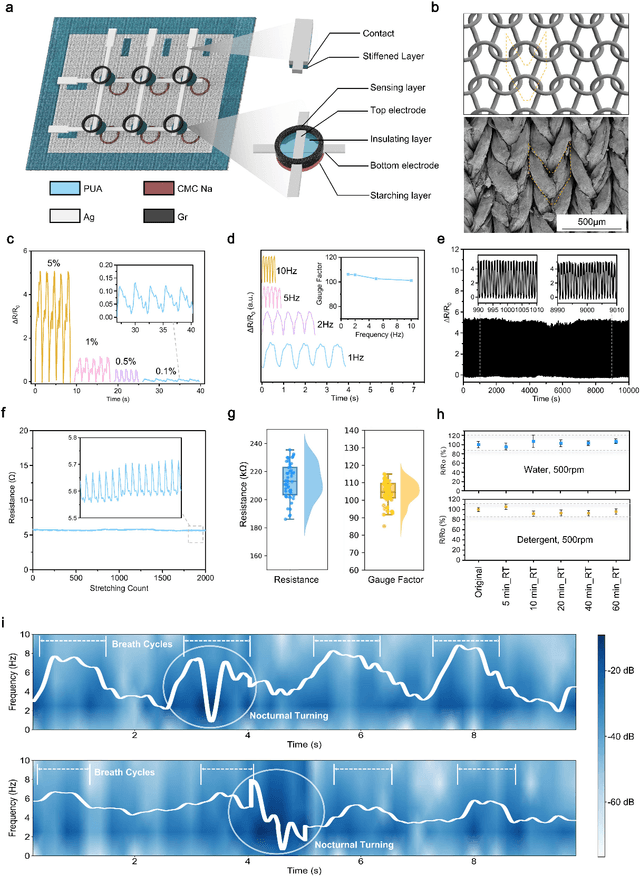
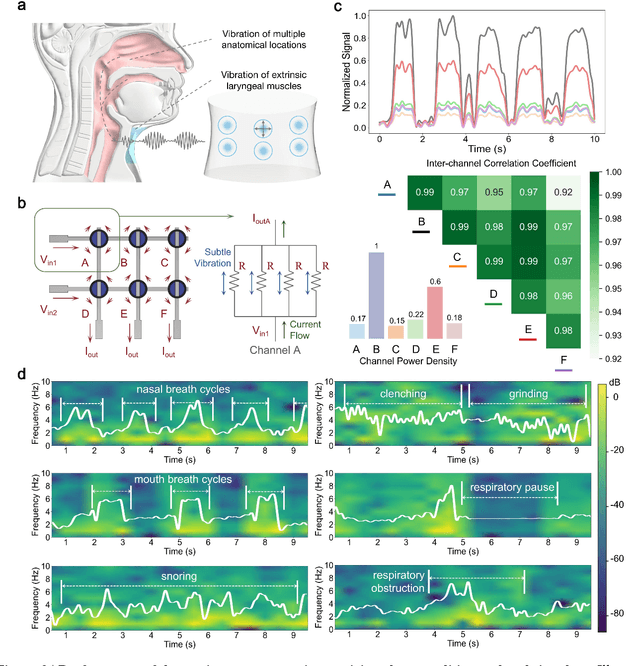
Abstract:Continuous monitoring and accurate detection of complex sleep patterns associated to different sleep-related conditions is essential, not only for enhancing sleep quality but also for preventing the risk of developing chronic illnesses associated to unhealthy sleep. Despite significant advances in research, achieving versatile recognition of various unhealthy and sub-healthy sleep patterns with simple wearable devices at home remains a significant challenge. Here, we report a robust and durable ultrasensitive strain sensor array printed on a smart garment, in its collar region. This solution allows detecting subtle vibrations associated with multiple sleep patterns at the extrinsic laryngeal muscles. Equipped with a deep learning neural network, it can precisely identify six sleep states-nasal breathing, mouth breathing, snoring, bruxism, central sleep apnea (CSA), and obstructive sleep apnea (OSA)-with an impressive accuracy of 98.6%, all without requiring specific positioning. We further demonstrate its explainability and generalization capabilities in practical applications. Explainable artificial intelligence (XAI) visualizations reflect comprehensive signal pattern analysis with low bias. Transfer learning tests show that the system can achieve high accuracy (overall accuracy of 95%) on new users with very few-shot learning (less than 15 samples per class). The scalable manufacturing process, robustness, high accuracy, and excellent generalization of the smart garment make it a promising tool for next-generation continuous sleep monitoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge