Yiping Liu
College of Computer Science and Electronic Engineering, Hunan University
Retrofit: Continual Learning with Bounded Forgetting for Security Applications
Nov 14, 2025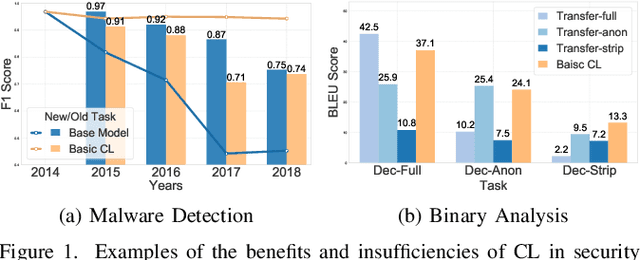
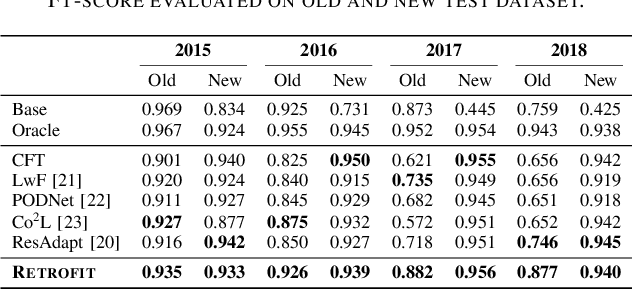
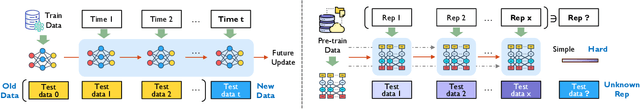
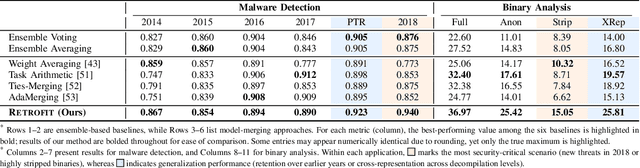
Abstract:Modern security analytics are increasingly powered by deep learning models, but their performance often degrades as threat landscapes evolve and data representations shift. While continual learning (CL) offers a promising paradigm to maintain model effectiveness, many approaches rely on full retraining or data replay, which are infeasible in data-sensitive environments. Moreover, existing methods remain inadequate for security-critical scenarios, facing two coupled challenges in knowledge transfer: preserving prior knowledge without old data and integrating new knowledge with minimal interference. We propose RETROFIT, a data retrospective-free continual learning method that achieves bounded forgetting for effective knowledge transfer. Our key idea is to consolidate previously trained and newly fine-tuned models, serving as teachers of old and new knowledge, through parameter-level merging that eliminates the need for historical data. To mitigate interference, we apply low-rank and sparse updates that confine parameter changes to independent subspaces, while a knowledge arbitration dynamically balances the teacher contributions guided by model confidence. Our evaluation on two representative applications demonstrates that RETROFIT consistently mitigates forgetting while maintaining adaptability. In malware detection under temporal drift, it substantially improves the retention score, from 20.2% to 38.6% over CL baselines, and exceeds the oracle upper bound on new data. In binary summarization across decompilation levels, where analyzing stripped binaries is especially challenging, RETROFIT achieves around twice the BLEU score of transfer learning used in prior work and surpasses all baselines in cross-representation generalization.
DynaQuant: Dynamic Mixed-Precision Quantization for Learned Image Compression
Nov 11, 2025Abstract:Prevailing quantization techniques in Learned Image Compression (LIC) typically employ a static, uniform bit-width across all layers, failing to adapt to the highly diverse data distributions and sensitivity characteristics inherent in LIC models. This leads to a suboptimal trade-off between performance and efficiency. In this paper, we introduce DynaQuant, a novel framework for dynamic mixed-precision quantization that operates on two complementary levels. First, we propose content-aware quantization, where learnable scaling and offset parameters dynamically adapt to the statistical variations of latent features. This fine-grained adaptation is trained end-to-end using a novel Distance-aware Gradient Modulator (DGM), which provides a more informative learning signal than the standard Straight-Through Estimator. Second, we introduce a data-driven, dynamic bit-width selector that learns to assign an optimal bit precision to each layer, dynamically reconfiguring the network's precision profile based on the input data. Our fully dynamic approach offers substantial flexibility in balancing rate-distortion (R-D) performance and computational cost. Experiments demonstrate that DynaQuant achieves rd performance comparable to full-precision models while significantly reducing computational and storage requirements, thereby enabling the practical deployment of advanced LIC on diverse hardware platforms.
ImageDDI: Image-enhanced Molecular Motif Sequence Representation for Drug-Drug Interaction Prediction
Aug 11, 2025Abstract:To mitigate the potential adverse health effects of simultaneous multi-drug use, including unexpected side effects and interactions, accurately identifying and predicting drug-drug interactions (DDIs) is considered a crucial task in the field of deep learning. Although existing methods have demonstrated promising performance, they suffer from the bottleneck of limited functional motif-based representation learning, as DDIs are fundamentally caused by motif interactions rather than the overall drug structures. In this paper, we propose an Image-enhanced molecular motif sequence representation framework for \textbf{DDI} prediction, called ImageDDI, which represents a pair of drugs from both global and local structures. Specifically, ImageDDI tokenizes molecules into functional motifs. To effectively represent a drug pair, their motifs are combined into a single sequence and embedded using a transformer-based encoder, starting from the local structure representation. By leveraging the associations between drug pairs, ImageDDI further enhances the spatial representation of molecules using global molecular image information (e.g. texture, shadow, color, and planar spatial relationships). To integrate molecular visual information into functional motif sequence, ImageDDI employs Adaptive Feature Fusion, enhancing the generalization of ImageDDI by dynamically adapting the fusion process of feature representations. Experimental results on widely used datasets demonstrate that ImageDDI outperforms state-of-the-art methods. Moreover, extensive experiments show that ImageDDI achieved competitive performance in both 2D and 3D image-enhanced scenarios compared to other models.
Kimi K2: Open Agentic Intelligence
Jul 28, 2025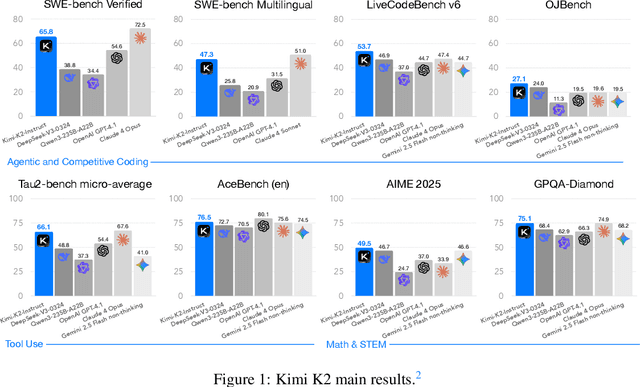

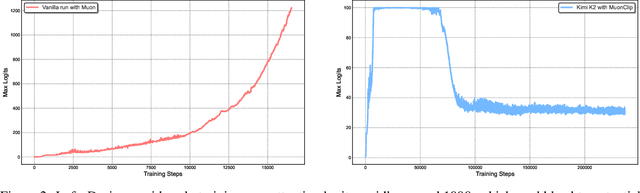
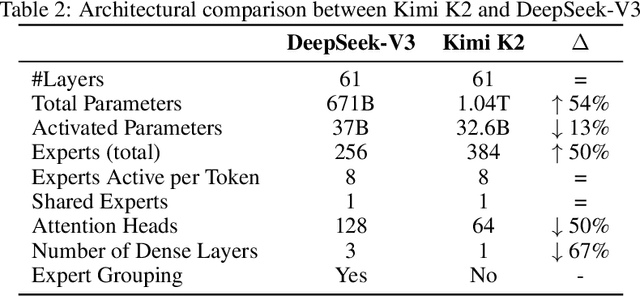
Abstract:We introduce Kimi K2, a Mixture-of-Experts (MoE) large language model with 32 billion activated parameters and 1 trillion total parameters. We propose the MuonClip optimizer, which improves upon Muon with a novel QK-clip technique to address training instability while enjoying the advanced token efficiency of Muon. Based on MuonClip, K2 was pre-trained on 15.5 trillion tokens with zero loss spike. During post-training, K2 undergoes a multi-stage post-training process, highlighted by a large-scale agentic data synthesis pipeline and a joint reinforcement learning (RL) stage, where the model improves its capabilities through interactions with real and synthetic environments. Kimi K2 achieves state-of-the-art performance among open-source non-thinking models, with strengths in agentic capabilities. Notably, K2 obtains 66.1 on Tau2-Bench, 76.5 on ACEBench (En), 65.8 on SWE-Bench Verified, and 47.3 on SWE-Bench Multilingual -- surpassing most open and closed-sourced baselines in non-thinking settings. It also exhibits strong capabilities in coding, mathematics, and reasoning tasks, with a score of 53.7 on LiveCodeBench v6, 49.5 on AIME 2025, 75.1 on GPQA-Diamond, and 27.1 on OJBench, all without extended thinking. These results position Kimi K2 as one of the most capable open-source large language models to date, particularly in software engineering and agentic tasks. We release our base and post-trained model checkpoints to facilitate future research and applications of agentic intelligence.
Dataset Distillation as Data Compression: A Rate-Utility Perspective
Jul 23, 2025Abstract:Driven by the ``scale-is-everything'' paradigm, modern machine learning increasingly demands ever-larger datasets and models, yielding prohibitive computational and storage requirements. Dataset distillation mitigates this by compressing an original dataset into a small set of synthetic samples, while preserving its full utility. Yet, existing methods either maximize performance under fixed storage budgets or pursue suitable synthetic data representations for redundancy removal, without jointly optimizing both objectives. In this work, we propose a joint rate-utility optimization method for dataset distillation. We parameterize synthetic samples as optimizable latent codes decoded by extremely lightweight networks. We estimate the Shannon entropy of quantized latents as the rate measure and plug any existing distillation loss as the utility measure, trading them off via a Lagrange multiplier. To enable fair, cross-method comparisons, we introduce bits per class (bpc), a precise storage metric that accounts for sample, label, and decoder parameter costs. On CIFAR-10, CIFAR-100, and ImageNet-128, our method achieves up to $170\times$ greater compression than standard distillation at comparable accuracy. Across diverse bpc budgets, distillation losses, and backbone architectures, our approach consistently establishes better rate-utility trade-offs.
A Reduction-Driven Local Search for the Generalized Independent Set Problem
May 27, 2025Abstract:The Generalized Independent Set (GIS) problem extends the classical maximum independent set problem by incorporating profits for vertices and penalties for edges. This generalized problem has been identified in diverse applications in fields such as forest harvest planning, competitive facility location, social network analysis, and even machine learning. However, solving the GIS problem in large-scale, real-world networks remains computationally challenging. In this paper, we explore data reduction techniques to address this challenge. We first propose 14 reduction rules that can reduce the input graph with rigorous optimality guarantees. We then present a reduction-driven local search (RLS) algorithm that integrates these reduction rules into the pre-processing, the initial solution generation, and the local search components in a computationally efficient way. The RLS is empirically evaluated on 278 graphs arising from different application scenarios. The results indicates that the RLS is highly competitive -- For most graphs, it achieves significantly superior solutions compared to other known solvers, and it effectively provides solutions for graphs exceeding 260 million edges, a task at which every other known method fails. Analysis also reveals that the data reduction plays a key role in achieving such a competitive performance.
AdaptMol: Adaptive Fusion from Sequence String to Topological Structure for Few-shot Drug Discovery
May 17, 2025



Abstract:Accurate molecular property prediction (MPP) is a critical step in modern drug development. However, the scarcity of experimental validation data poses a significant challenge to AI-driven research paradigms. Under few-shot learning scenarios, the quality of molecular representations directly dictates the theoretical upper limit of model performance. We present AdaptMol, a prototypical network integrating Adaptive multimodal fusion for Molecular representation. This framework employs a dual-level attention mechanism to dynamically integrate global and local molecular features derived from two modalities: SMILES sequences and molecular graphs. (1) At the local level, structural features such as atomic interactions and substructures are extracted from molecular graphs, emphasizing fine-grained topological information; (2) At the global level, the SMILES sequence provides a holistic representation of the molecule. To validate the necessity of multimodal adaptive fusion, we propose an interpretable approach based on identifying molecular active substructures to demonstrate that multimodal adaptive fusion can efficiently represent molecules. Extensive experiments on three commonly used benchmarks under 5-shot and 10-shot settings demonstrate that AdaptMol achieves state-of-the-art performance in most cases. The rationale-extracted method guides the fusion of two modalities and highlights the importance of both modalities.
Data Pricing for Graph Neural Networks without Pre-purchased Inspection
Feb 12, 2025



Abstract:Machine learning (ML) models have become essential tools in various scenarios. Their effectiveness, however, hinges on a substantial volume of data for satisfactory performance. Model marketplaces have thus emerged as crucial platforms bridging model consumers seeking ML solutions and data owners possessing valuable data. These marketplaces leverage model trading mechanisms to properly incentive data owners to contribute their data, and return a well performing ML model to the model consumers. However, existing model trading mechanisms often assume the data owners are willing to share their data before being paid, which is not reasonable in real world. Given that, we propose a novel mechanism, named Structural Importance based Model Trading (SIMT) mechanism, that assesses the data importance and compensates data owners accordingly without disclosing the data. Specifically, SIMT procures feature and label data from data owners according to their structural importance, and then trains a graph neural network for model consumers. Theoretically, SIMT ensures incentive compatible, individual rational and budget feasible. The experiments on five popular datasets validate that SIMT consistently outperforms vanilla baselines by up to $40\%$ in both MacroF1 and MicroF1.
Efficient Top-k s-Biplexes Search over Large Bipartite Graphs
Sep 27, 2024Abstract:In a bipartite graph, a subgraph is an $s$-biplex if each vertex of the subgraph is adjacent to all but at most $s$ vertices on the opposite set. The enumeration of $s$-biplexes from a given graph is a fundamental problem in bipartite graph analysis. However, in real-world data engineering, finding all $s$-biplexes is neither necessary nor computationally affordable. A more realistic problem is to identify some of the largest $s$-biplexes from the large input graph. We formulate the problem as the {\em top-$k$ $s$-biplex search (TBS) problem}, which aims to find the top-$k$ maximal $s$-biplexes with the most vertices, where $k$ is an input parameter. We prove that the TBS problem is NP-hard for any fixed $k\ge 1$. Then, we propose a branching algorithm, named MVBP, that breaks the simple $2^n$ enumeration algorithm. Furthermore, from a practical perspective, we investigate three techniques to improve the performance of MVBP: 2-hop decomposition, single-side bounds, and progressive search. Complexity analysis shows that the improved algorithm, named FastMVBP, has a running time $O^*(\gamma_s^{d_2})$, where $\gamma_s<2$, and $d_2$ is a parameter much smaller than the number of vertex in the sparse real-world graphs, e.g. $d_2$ is only $67$ in the AmazonRatings dataset which has more than $3$ million vertices. Finally, we conducted extensive experiments on eight real-world and synthetic datasets to demonstrate the empirical efficiency of the proposed algorithms. In particular, FastMVBP outperforms the benchmark algorithms by up to three orders of magnitude in several instances.
MoFormer: Multi-objective Antimicrobial Peptide Generation Based on Conditional Transformer Joint Multi-modal Fusion Descriptor
Jun 03, 2024



Abstract:Deep learning holds a big promise for optimizing existing peptides with more desirable properties, a critical step towards accelerating new drug discovery. Despite the recent emergence of several optimized Antimicrobial peptides(AMP) generation methods, multi-objective optimizations remain still quite challenging for the idealism-realism tradeoff. Here, we establish a multi-objective AMP synthesis pipeline (MoFormer) for the simultaneous optimization of multi-attributes of AMPs. MoFormer improves the desired attributes of AMP sequences in a highly structured latent space, guided by conditional constraints and fine-grained multi-descriptor.We show that MoFormer outperforms existing methods in the generation task of enhanced antimicrobial activity and minimal hemolysis. We also utilize a Pareto-based non-dominated sorting algorithm and proxies based on large model fine-tuning to hierarchically rank the candidates. We demonstrate substantial property improvement using MoFormer from two perspectives: (1) employing molecular simulations and scoring interactions among amino acids to decipher the structure and functionality of AMPs; (2) visualizing latent space to examine the qualities and distribution features, verifying an effective means to facilitate multi-objective optimization AMPs with design constraints
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge