Sen Song
Signal-Adaptive Trust Regions for Gradient-Free Optimization of Recurrent Spiking Neural Networks
Jan 29, 2026Abstract:Recurrent spiking neural networks (RSNNs) are a promising substrate for energy-efficient control policies, but training them for high-dimensional, long-horizon reinforcement learning remains challenging. Population-based, gradient-free optimization circumvents backpropagation through non-differentiable spike dynamics by estimating gradients. However, with finite populations, high variance of these estimates can induce harmful and overly aggressive update steps. Inspired by trust-region methods in reinforcement learning that constrain policy updates in distribution space, we propose \textbf{Signal-Adaptive Trust Regions (SATR)}, a distributional update rule that constrains relative change by bounding KL divergence normalized by an estimated signal energy. SATR automatically expands the trust region under strong signals and contracts it when updates are noise-dominated. We instantiate SATR for Bernoulli connectivity distributions, which have shown strong empirical performance for RSNN optimization. Across a suite of high-dimensional continuous-control benchmarks, SATR improves stability under limited populations and reaches competitive returns against strong baselines including PPO-LSTM. In addition, to make SATR practical at scale, we introduce a bitset implementation for binary spiking and binary weights, substantially reducing wall-clock training time and enabling fast RSNN policy search.
Improve Decoding Factuality by Token-wise Cross Layer Entropy of Large Language Models
Feb 05, 2025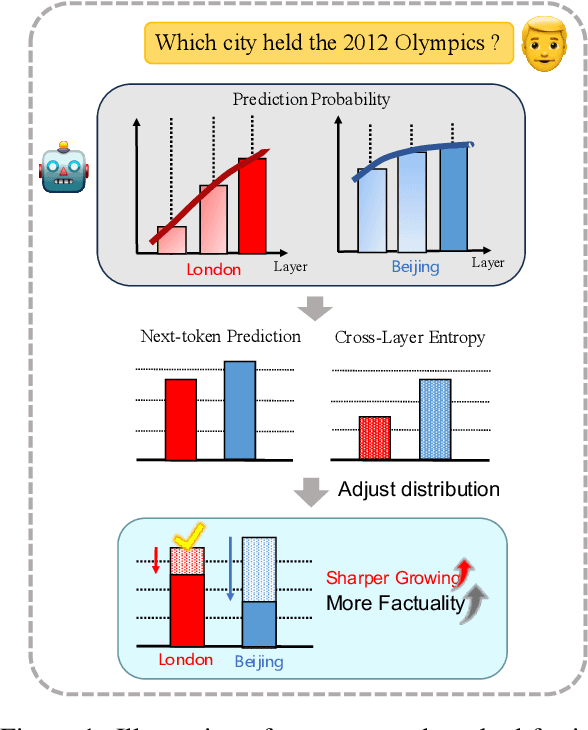
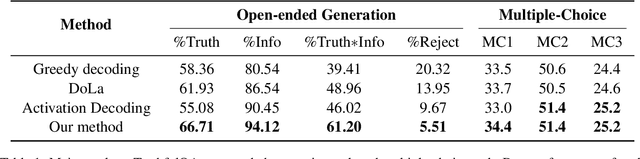
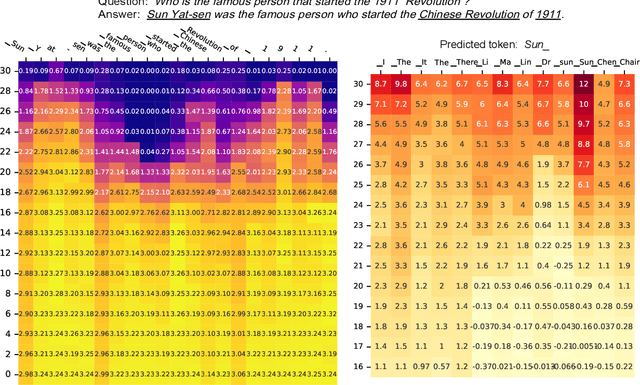
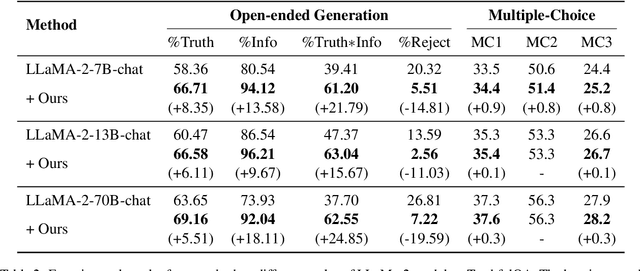
Abstract:Despite their impressive capacities, Large language models (LLMs) often struggle with the hallucination issue of generating inaccurate or fabricated content even when they possess correct knowledge. In this paper, we extend the exploration of the correlation between hidden-state prediction changes and output factuality into a deeper, token-wise level. Based on the insights , we propose cross-layer Entropy eNhanced Decoding (END), a decoding method that mitigates hallucinations without requiring extra training. END leverages inner probability changes across layers to individually quantify the factual knowledge required for each candidate token, and adjusts the final predicting distribution to prioritize tokens with higher factuality. Experiments on both hallucination and QA benchmarks demonstrate that END significantly enhances the truthfulness and informativeness of generated content while maintaining robust QA accuracy. Moreover, our work provides a deeper perspective on understanding the correlations between inherent knowledge and output factuality.
Dynamic-Attention-based EEG State Transition Modeling for Emotion Recognition
Nov 07, 2024



Abstract:Electroencephalogram (EEG)-based emotion decoding can objectively quantify people's emotional state and has broad application prospects in human-computer interaction and early detection of emotional disorders. Recently emerging deep learning architectures have significantly improved the performance of EEG emotion decoding. However, existing methods still fall short of fully capturing the complex spatiotemporal dynamics of neural signals, which are crucial for representing emotion processing. This study proposes a Dynamic-Attention-based EEG State Transition (DAEST) modeling method to characterize EEG spatiotemporal dynamics. The model extracts spatiotemporal components of EEG that represent multiple parallel neural processes and estimates dynamic attention weights on these components to capture transitions in brain states. The model is optimized within a contrastive learning framework for cross-subject emotion recognition. The proposed method achieved state-of-the-art performance on three publicly available datasets: FACED, SEED, and SEED-V. It achieved 75.4% accuracy in the binary classification of positive and negative emotions and 59.3% in nine-class discrete emotion classification on the FACED dataset, 88.1% in the three-class classification of positive, negative, and neutral emotions on the SEED dataset, and 73.6% in five-class discrete emotion classification on the SEED-V dataset. The learned EEG spatiotemporal patterns and dynamic transition properties offer valuable insights into neural dynamics underlying emotion processing.
Contrastive Learning of Shared Spatiotemporal EEG Representations Across Individuals for Naturalistic Neuroscience
Feb 22, 2024



Abstract:Neural representations induced by naturalistic stimuli offer insights into how humans respond to peripheral stimuli in daily life. The key to understanding the general neural mechanisms underlying naturalistic stimuli processing involves aligning neural activities across individuals and extracting inter-subject shared neural representations. Targeting the Electroencephalogram (EEG) technique, known for its rich spatial and temporal information, this study presents a general framework for Contrastive Learning of Shared SpatioTemporal EEG Representations across individuals (CL-SSTER). Harnessing the representational capabilities of contrastive learning, CL-SSTER utilizes a neural network to maximize the similarity of EEG representations across individuals for identical stimuli, contrasting with those for varied stimuli. The network employed spatial and temporal convolutions to simultaneously learn the spatial and temporal patterns inherent in EEG. The versatility of CL-SSTER was demonstrated on three EEG datasets, including a synthetic dataset, a speech audio EEG dataset, and an emotional video EEG dataset. CL-SSTER attained the highest inter-subject correlation (ISC) values compared to the state-of-the-art ISC methods. The latent representations generated by CL-SSTER exhibited reliable spatiotemporal EEG patterns, which can be explained by specific aspects of the stimuli. CL-SSTER serves as an interpretable and scalable foundational framework for the identification of inter-subject shared neural representations in the realm of naturalistic neuroscience.
UniMem: Towards a Unified View of Long-Context Large Language Models
Feb 05, 2024



Abstract:Long-context processing is a critical ability that constrains the applicability of large language models. Although there exist various methods devoted to enhancing the long-context processing ability of large language models (LLMs), they are developed in an isolated manner and lack systematic analysis and integration of their strengths, hindering further developments. In this paper, we introduce UniMem, a unified framework that reformulates existing long-context methods from the view of memory augmentation of LLMs. UniMem is characterized by four key dimensions: Memory Management, Memory Writing, Memory Reading, and Memory Injection, providing a systematic theory for understanding various long-context methods. We reformulate 16 existing methods based on UniMem and analyze four representative methods: Transformer-XL, Memorizing Transformer, RMT, and Longformer into equivalent UniMem forms to reveal their design principles and strengths. Based on these analyses, we propose UniMix, an innovative approach that integrates the strengths of these algorithms. Experimental results show that UniMix achieves superior performance in handling long contexts with significantly lower perplexity than baselines.
Brain-Like Replay Naturally Emerges in Reinforcement Learning Agents
Feb 02, 2024Abstract:Can replay, as a widely observed neural activity pattern in brain regions, particularly in the hippocampus and neocortex, emerge in an artificial agent? If yes, does it contribute to the tasks? In this work, without heavy dependence on complex assumptions, we discover naturally emergent replay under task-optimized paradigm using a recurrent neural network-based reinforcement learning model, which mimics the hippocampus and prefrontal cortex, as well as their intercommunication and the sensory cortex input. The emergent replay in the hippocampus, which results from the episodic memory and cognitive map as well as environment observations, well resembles animal experimental data and serves as an effective indicator of high task performance. The model also successfully reproduces local and nonlocal replay, which matches the human experimental data. Our work provides a new avenue for understanding the mechanisms behind replay.
OpenChat: Advancing Open-source Language Models with Mixed-Quality Data
Sep 20, 2023



Abstract:Nowadays, open-source large language models like LLaMA have emerged. Recent developments have incorporated supervised fine-tuning (SFT) and reinforcement learning fine-tuning (RLFT) to align these models with human goals. However, SFT methods treat all training data with mixed quality equally, while RLFT methods require high-quality pairwise or ranking-based preference data. In this study, we present a novel framework, named OpenChat, to advance open-source language models with mixed-quality data. Specifically, we consider the general SFT training data, consisting of a small amount of expert data mixed with a large proportion of sub-optimal data, without any preference labels. We propose the C(onditioned)-RLFT, which regards different data sources as coarse-grained reward labels and learns a class-conditioned policy to leverage complementary data quality information. Interestingly, the optimal policy in C-RLFT can be easily solved through single-stage, RL-free supervised learning, which is lightweight and avoids costly human preference labeling. Through extensive experiments on three standard benchmarks, our openchat-13b fine-tuned with C-RLFT achieves the highest average performance among all 13b open-source language models. Moreover, we use AGIEval to validate the model generalization performance, in which only openchat-13b surpasses the base model. Finally, we conduct a series of analyses to shed light on the effectiveness and robustness of OpenChat. Our code, data, and models are publicly available at https://github.com/imoneoi/openchat.
Evolving Connectivity for Recurrent Spiking Neural Networks
May 28, 2023



Abstract:Recurrent spiking neural networks (RSNNs) hold great potential for advancing artificial general intelligence, as they draw inspiration from the biological nervous system and show promise in modeling complex dynamics. However, the widely-used surrogate gradient-based training methods for RSNNs are inherently inaccurate and unfriendly to neuromorphic hardware. To address these limitations, we propose the evolving connectivity (EC) framework, an inference-only method for training RSNNs. The EC framework reformulates weight-tuning as a search into parameterized connection probability distributions, and employs Natural Evolution Strategies (NES) for optimizing these distributions. Our EC framework circumvents the need for gradients and features hardware-friendly characteristics, including sparse boolean connections and high scalability. We evaluate EC on a series of standard robotic locomotion tasks, where it achieves comparable performance with deep neural networks and outperforms gradient-trained RSNNs, even solving the complex 17-DoF humanoid task. Additionally, the EC framework demonstrates a two to three fold speedup in efficiency compared to directly evolving parameters. By providing a performant and hardware-friendly alternative, the EC framework lays the groundwork for further energy-efficient applications of RSNNs and advances the development of neuromorphic devices.
Local Hypergraph-based Nested Named Entity Recognition as Query-based Sequence Labeling
May 04, 2022



Abstract:There has been a growing academic interest in the recognition of nested named entities in many domains. We tackle the task with a novel local hypergraph-based method: We first propose start token candidates and generate corresponding queries with their surrounding context, then use a query-based sequence labeling module to form a local hypergraph for each candidate. An end token estimator is used to correct the hypergraphs and get the final predictions. Compared to span-based approaches, our method is free of the high computation cost of span sampling and the risk of losing long entities. Sequential prediction makes it easier to leverage information in word order inside nested structures, and richer representations are built with a local hypergraph. Experiments show that our proposed method outperforms all the previous hypergraph-based and sequence labeling approaches with large margins on all four nested datasets. It achieves a new state-of-the-art F1 score on the ACE 2004 dataset and competitive F1 scores with previous state-of-the-art methods on three other nested NER datasets: ACE 2005, GENIA, and KBP 2017.
Simulated annealing for optimization of graphs and sequences
Oct 01, 2021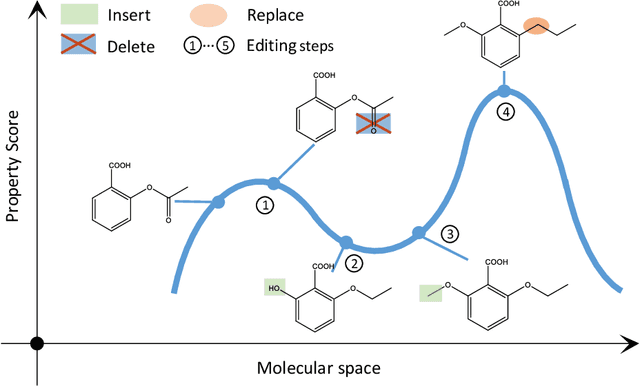
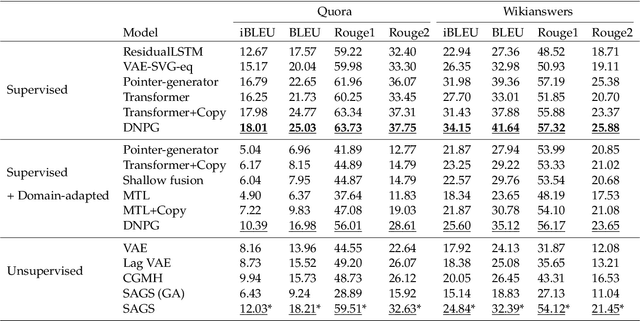
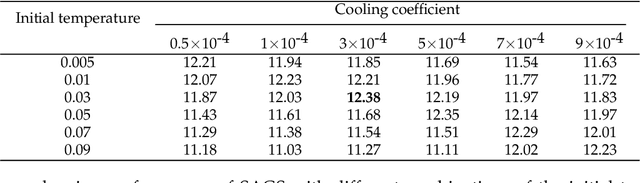
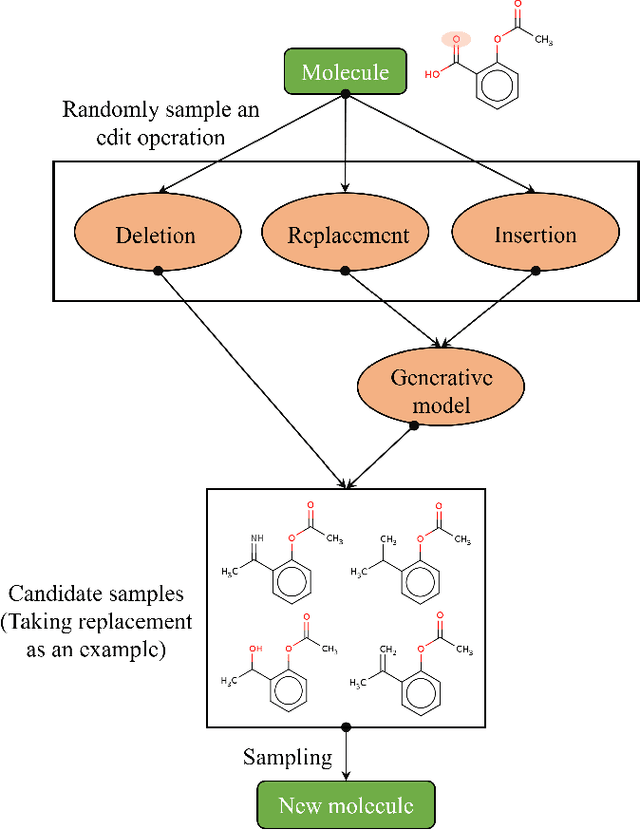
Abstract:Optimization of discrete structures aims at generating a new structure with the better property given an existing one, which is a fundamental problem in machine learning. Different from the continuous optimization, the realistic applications of discrete optimization (e.g., text generation) are very challenging due to the complex and long-range constraints, including both syntax and semantics, in discrete structures. In this work, we present SAGS, a novel Simulated Annealing framework for Graph and Sequence optimization. The key idea is to integrate powerful neural networks into metaheuristics (e.g., simulated annealing, SA) to restrict the search space in discrete optimization. We start by defining a sophisticated objective function, involving the property of interest and pre-defined constraints (e.g., grammar validity). SAGS searches from the discrete space towards this objective by performing a sequence of local edits, where deep generative neural networks propose the editing content and thus can control the quality of editing. We evaluate SAGS on paraphrase generation and molecule generation for sequence optimization and graph optimization, respectively. Extensive results show that our approach achieves state-of-the-art performance compared with existing paraphrase generation methods in terms of both automatic and human evaluations. Further, SAGS also significantly outperforms all the previous methods in molecule generation.
* This article is an accepted manuscript of Neurocomputing. arXiv admin note: substantial text overlap with arXiv:1909.03588
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge