Xianggen Liu
DrugLLM: Open Large Language Model for Few-shot Molecule Generation
May 07, 2024



Abstract:Large Language Models (LLMs) have made great strides in areas such as language processing and computer vision. Despite the emergence of diverse techniques to improve few-shot learning capacity, current LLMs fall short in handling the languages in biology and chemistry. For example, they are struggling to capture the relationship between molecule structure and pharmacochemical properties. Consequently, the few-shot learning capacity of small-molecule drug modification remains impeded. In this work, we introduced DrugLLM, a LLM tailored for drug design. During the training process, we employed Group-based Molecular Representation (GMR) to represent molecules, arranging them in sequences that reflect modifications aimed at enhancing specific molecular properties. DrugLLM learns how to modify molecules in drug discovery by predicting the next molecule based on past modifications. Extensive computational experiments demonstrate that DrugLLM can generate new molecules with expected properties based on limited examples, presenting a powerful few-shot molecule generation capacity.
MSGNet: Learning Multi-Scale Inter-Series Correlations for Multivariate Time Series Forecasting
Dec 31, 2023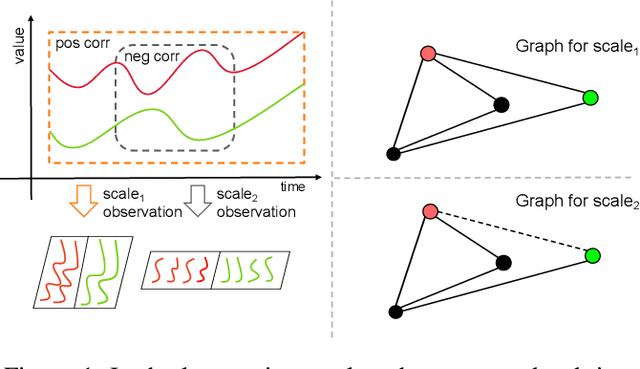
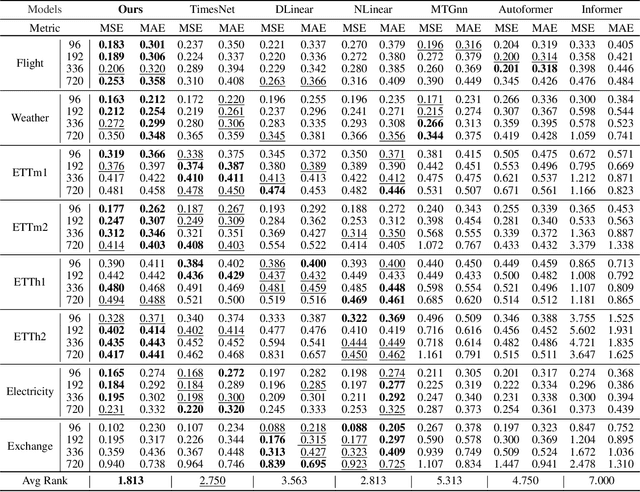
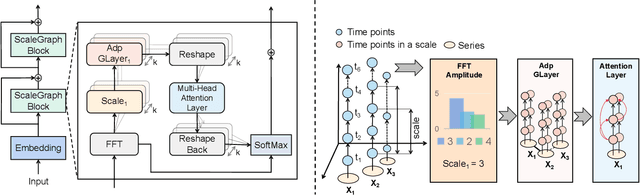
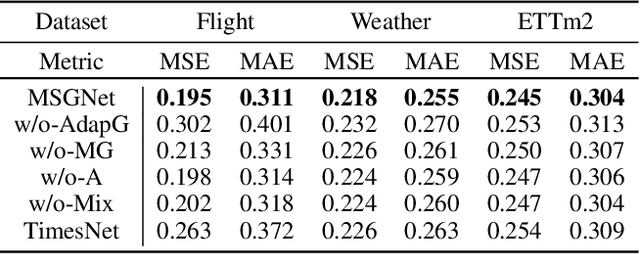
Abstract:Multivariate time series forecasting poses an ongoing challenge across various disciplines. Time series data often exhibit diverse intra-series and inter-series correlations, contributing to intricate and interwoven dependencies that have been the focus of numerous studies. Nevertheless, a significant research gap remains in comprehending the varying inter-series correlations across different time scales among multiple time series, an area that has received limited attention in the literature. To bridge this gap, this paper introduces MSGNet, an advanced deep learning model designed to capture the varying inter-series correlations across multiple time scales using frequency domain analysis and adaptive graph convolution. By leveraging frequency domain analysis, MSGNet effectively extracts salient periodic patterns and decomposes the time series into distinct time scales. The model incorporates a self-attention mechanism to capture intra-series dependencies, while introducing an adaptive mixhop graph convolution layer to autonomously learn diverse inter-series correlations within each time scale. Extensive experiments are conducted on several real-world datasets to showcase the effectiveness of MSGNet. Furthermore, MSGNet possesses the ability to automatically learn explainable multi-scale inter-series correlations, exhibiting strong generalization capabilities even when applied to out-of-distribution samples.
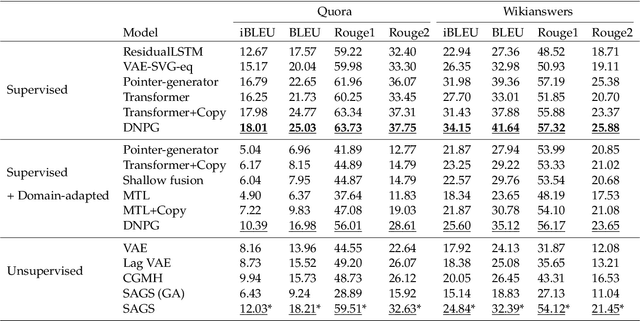
Vector-Quantized Prompt Learning for Paraphrase Generation
Nov 25, 2023

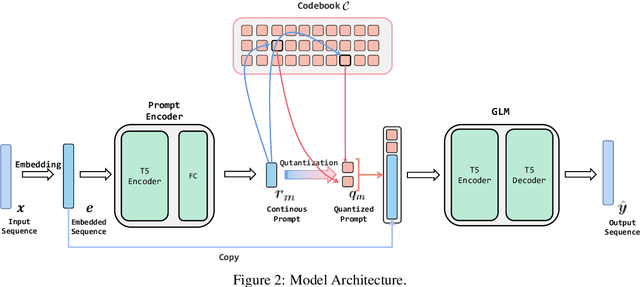

Abstract:Deep generative modeling of natural languages has achieved many successes, such as producing fluent sentences and translating from one language into another. However, the development of generative modeling techniques for paraphrase generation still lags behind largely due to the challenges in addressing the complex conflicts between expression diversity and semantic preservation. This paper proposes to generate diverse and high-quality paraphrases by exploiting the pre-trained models with instance-dependent prompts. To learn generalizable prompts, we assume that the number of abstract transforming patterns of paraphrase generation (governed by prompts) is finite and usually not large. Therefore, we present vector-quantized prompts as the cues to control the generation of pre-trained models. Extensive experiments demonstrate that the proposed method achieves new state-of-art results on three benchmark datasets, including Quora, Wikianswers, and MSCOCO. We will release all the code upon acceptance.
TimeSQL: Improving Multivariate Time Series Forecasting with Multi-Scale Patching and Smooth Quadratic Loss
Nov 19, 2023Abstract:Time series is a special type of sequence data, a sequence of real-valued random variables collected at even intervals of time. The real-world multivariate time series comes with noises and contains complicated local and global temporal dynamics, making it difficult to forecast the future time series given the historical observations. This work proposes a simple and effective framework, coined as TimeSQL, which leverages multi-scale patching and smooth quadratic loss (SQL) to tackle the above challenges. The multi-scale patching transforms the time series into two-dimensional patches with different length scales, facilitating the perception of both locality and long-term correlations in time series. SQL is derived from the rational quadratic kernel and can dynamically adjust the gradients to avoid overfitting to the noises and outliers. Theoretical analysis demonstrates that, under mild conditions, the effect of the noises on the model with SQL is always smaller than that with MSE. Based on the two modules, TimeSQL achieves new state-of-the-art performance on the eight real-world benchmark datasets. Further ablation studies indicate that the key modules in TimeSQL could also enhance the results of other models for multivariate time series forecasting, standing as plug-and-play techniques.
Weakly Supervised Reasoning by Neuro-Symbolic Approaches
Sep 19, 2023



Abstract:Deep learning has largely improved the performance of various natural language processing (NLP) tasks. However, most deep learning models are black-box machinery, and lack explicit interpretation. In this chapter, we will introduce our recent progress on neuro-symbolic approaches to NLP, which combines different schools of AI, namely, symbolism and connectionism. Generally, we will design a neural system with symbolic latent structures for an NLP task, and apply reinforcement learning or its relaxation to perform weakly supervised reasoning in the downstream task. Our framework has been successfully applied to various tasks, including table query reasoning, syntactic structure reasoning, information extraction reasoning, and rule reasoning. For each application, we will introduce the background, our approach, and experimental results.
GPT-NAS: Neural Architecture Search with the Generative Pre-Trained Model
May 09, 2023Abstract:Neural Architecture Search (NAS) has emerged as one of the effective methods to design the optimal neural network architecture automatically. Although neural architectures have achieved human-level performances in several tasks, few of them are obtained from the NAS method. The main reason is the huge search space of neural architectures, making NAS algorithms inefficient. This work presents a novel architecture search algorithm, called GPT-NAS, that optimizes neural architectures by Generative Pre-Trained (GPT) model. In GPT-NAS, we assume that a generative model pre-trained on a large-scale corpus could learn the fundamental law of building neural architectures. Therefore, GPT-NAS leverages the generative pre-trained (GPT) model to propose reasonable architecture components given the basic one. Such an approach can largely reduce the search space by introducing prior knowledge in the search process. Extensive experimental results show that our GPT-NAS method significantly outperforms seven manually designed neural architectures and thirteen architectures provided by competing NAS methods. In addition, our ablation study indicates that the proposed algorithm improves the performance of finely tuned neural architectures by up to about 12% compared to those without GPT, further demonstrating its effectiveness in searching neural architectures.
Pairwise Half-graph Discrimination: A Simple Graph-level Self-supervised Strategy for Pre-training Graph Neural Networks
Oct 26, 2021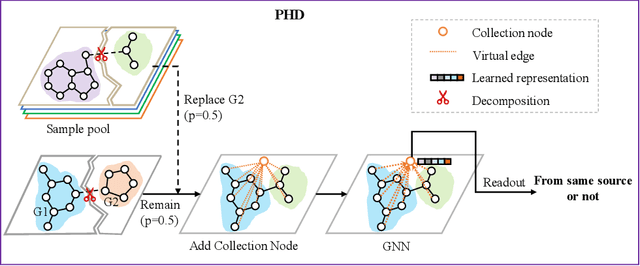
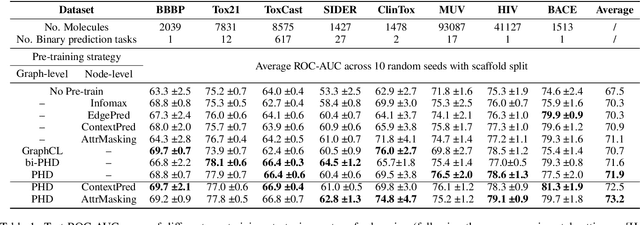
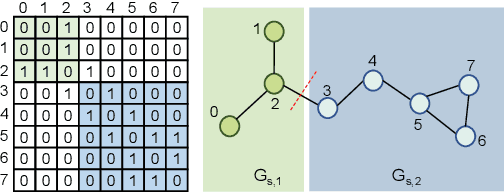

Abstract:Self-supervised learning has gradually emerged as a powerful technique for graph representation learning. However, transferable, generalizable, and robust representation learning on graph data still remains a challenge for pre-training graph neural networks. In this paper, we propose a simple and effective self-supervised pre-training strategy, named Pairwise Half-graph Discrimination (PHD), that explicitly pre-trains a graph neural network at graph-level. PHD is designed as a simple binary classification task to discriminate whether two half-graphs come from the same source. Experiments demonstrate that the PHD is an effective pre-training strategy that offers comparable or superior performance on 13 graph classification tasks compared with state-of-the-art strategies, and achieves notable improvements when combined with node-level strategies. Moreover, the visualization of learned representation revealed that PHD strategy indeed empowers the model to learn graph-level knowledge like the molecular scaffold. These results have established PHD as a powerful and effective self-supervised learning strategy in graph-level representation learning.
Simulated annealing for optimization of graphs and sequences
Oct 01, 2021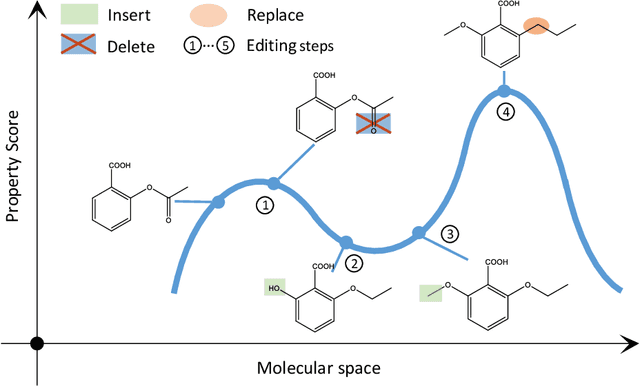
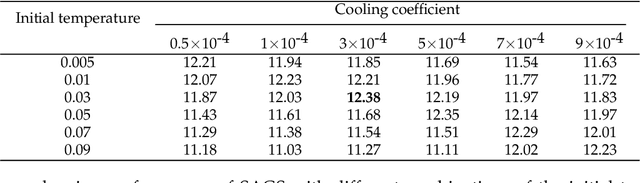
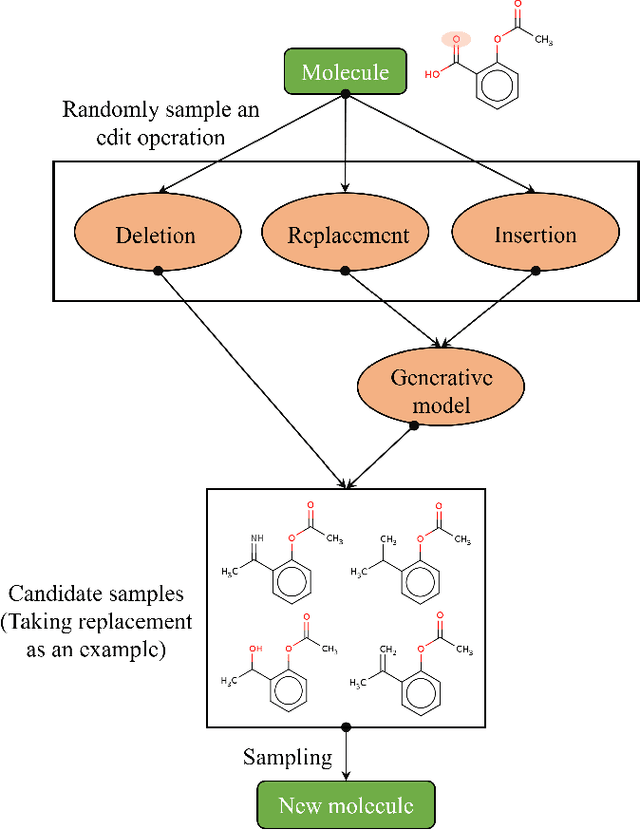
Abstract:Optimization of discrete structures aims at generating a new structure with the better property given an existing one, which is a fundamental problem in machine learning. Different from the continuous optimization, the realistic applications of discrete optimization (e.g., text generation) are very challenging due to the complex and long-range constraints, including both syntax and semantics, in discrete structures. In this work, we present SAGS, a novel Simulated Annealing framework for Graph and Sequence optimization. The key idea is to integrate powerful neural networks into metaheuristics (e.g., simulated annealing, SA) to restrict the search space in discrete optimization. We start by defining a sophisticated objective function, involving the property of interest and pre-defined constraints (e.g., grammar validity). SAGS searches from the discrete space towards this objective by performing a sequence of local edits, where deep generative neural networks propose the editing content and thus can control the quality of editing. We evaluate SAGS on paraphrase generation and molecule generation for sequence optimization and graph optimization, respectively. Extensive results show that our approach achieves state-of-the-art performance compared with existing paraphrase generation methods in terms of both automatic and human evaluations. Further, SAGS also significantly outperforms all the previous methods in molecule generation.
* This article is an accepted manuscript of Neurocomputing. arXiv admin note: substantial text overlap with arXiv:1909.03588
Contrastive Learning of Subject-Invariant EEG Representations for Cross-Subject Emotion Recognition
Sep 20, 2021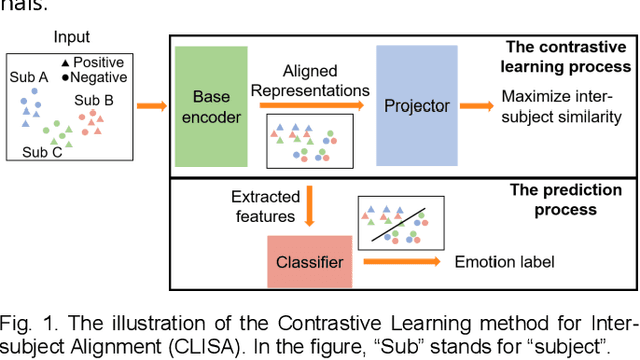
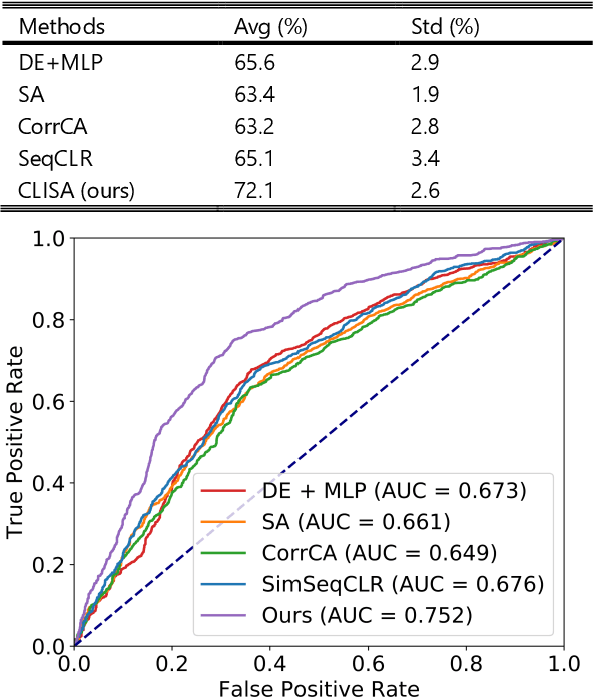
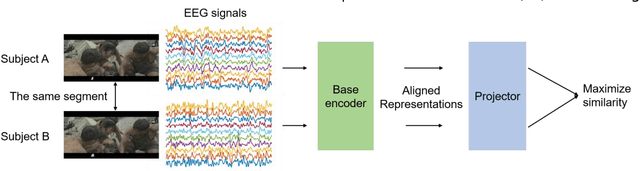
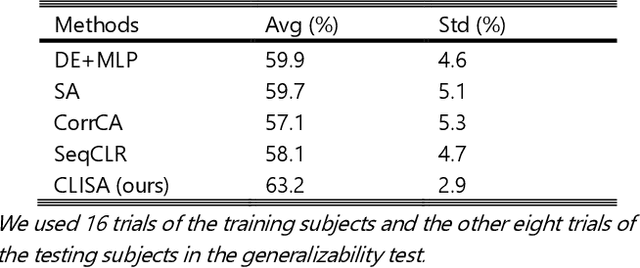
Abstract:Emotion recognition plays a vital role in human-machine interactions and daily healthcare. EEG signals have been reported to be informative and reliable for emotion recognition in recent years. However, the inter-subject variability of emotion-related EEG signals poses a great challenge for the practical use of EEG-based emotion recognition. Inspired by the recent neuroscience studies on inter-subject correlation, we proposed a Contrastive Learning method for Inter-Subject Alignment (CLISA) for reliable cross-subject emotion recognition. Contrastive learning was employed to minimize the inter-subject differences by maximizing the similarity in EEG signals across subjects when they received the same stimuli in contrast to different ones. Specifically, a convolutional neural network with depthwise spatial convolution and temporal convolution layers was applied to learn inter-subject aligned spatiotemporal representations from raw EEG signals. Then the aligned representations were used to extract differential entropy features for emotion classification. The performance of the proposed method was evaluated on our THU-EP dataset with 80 subjects and the publicly available SEED dataset with 15 subjects. Comparable or better cross-subject emotion recognition accuracy (i.e., 72.1% and 47.0% for binary and nine-class classification, respectively, on the THU-EP dataset and 86.3% on the SEED dataset for three-class classification) was achieved as compared to the state-of-the-art methods. The proposed method could be generalized well to unseen emotional stimuli as well. The CLISA method is therefore expected to considerably increase the practicality of EEG-based emotion recognition by operating in a "plug-and-play" manner. Furthermore, the learned spatiotemporal representations by CLISA could provide insights into the neural mechanisms of human emotion processing.
Pre-training of Graph Neural Network for Modeling Effects of Mutations on Protein-Protein Binding Affinity
Aug 28, 2020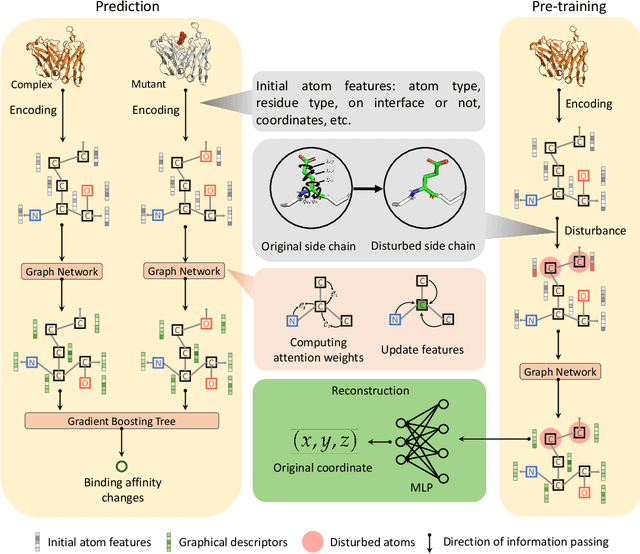
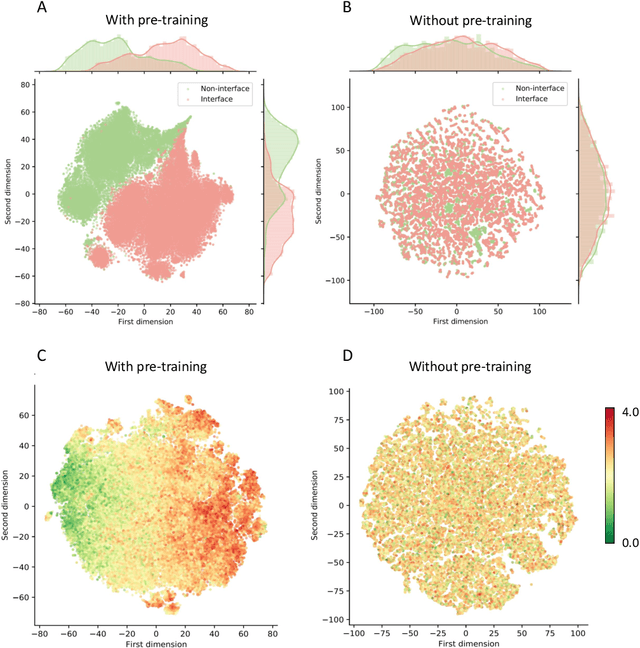
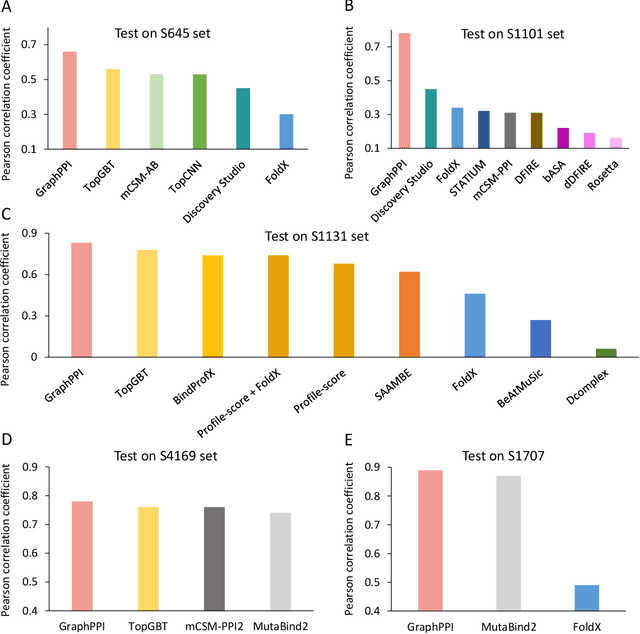
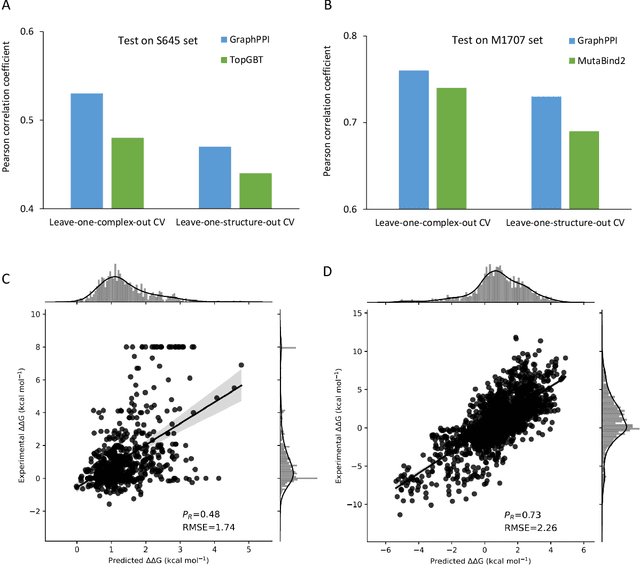
Abstract:Modeling the effects of mutations on the binding affinity plays a crucial role in protein engineering and drug design. In this study, we develop a novel deep learning based framework, named GraphPPI, to predict the binding affinity changes upon mutations based on the features provided by a graph neural network (GNN). In particular, GraphPPI first employs a well-designed pre-training scheme to enforce the GNN to capture the features that are predictive of the effects of mutations on binding affinity in an unsupervised manner and then integrates these graphical features with gradient-boosting trees to perform the prediction. Experiments showed that, without any annotated signals, GraphPPI can capture meaningful patterns of the protein structures. Also, GraphPPI achieved new state-of-the-art performance in predicting the binding affinity changes upon both single- and multi-point mutations on five benchmark datasets. In-depth analyses also showed GraphPPI can accurately estimate the effects of mutations on the binding affinity between SARS-CoV-2 and its neutralizing antibodies. These results have established GraphPPI as a powerful and useful computational tool in the studies of protein design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge