Lin Yao
JTok: On Token Embedding as another Axis of Scaling Law via Joint Token Self-modulation
Jan 31, 2026Abstract:LLMs have traditionally scaled along dense dimensions, where performance is coupled with near-linear increases in computational cost. While MoE decouples capacity from compute, it introduces large memory overhead and hardware efficiency challenges. To overcome these, we propose token-indexed parameters as a novel, orthogonal scaling axis that decouple model capacity from FLOPs. Specifically, we introduce Joint-Token (JTok) and Mixture of Joint-Token (JTok-M), which augment Transformer layers with modulation vectors retrieved from auxiliary embedding tables. These vectors modulate the backbone via lightweight, element-wise operations, incurring negligible FLOPs overhead. Extensive experiments on both dense and MoE backbones, spanning from 650M (190M + 460M embedding) to 61B (17B + 44B embedding) total parameters, demonstrate that our approach consistently reduces validation loss and significantly improves downstream task performance (e.g., +4.1 on MMLU, +8.3 on ARC, +8.9 on CEval). Rigorous isoFLOPs analysis further confirms that JTok-M fundamentally shifts the quality-compute Pareto frontier, achieving comparable model quality with 35% less compute relative to vanilla MoE architectures, and we validate that token-indexed parameters exhibit a predictable power-law scaling behavior. Moreover, our efficient implementation ensures that the overhead introduced by JTok and JTok-M remains marginal.
Innovator-VL: A Multimodal Large Language Model for Scientific Discovery
Jan 27, 2026Abstract:We present Innovator-VL, a scientific multimodal large language model designed to advance understanding and reasoning across diverse scientific domains while maintaining excellent performance on general vision tasks. Contrary to the trend of relying on massive domain-specific pretraining and opaque pipelines, our work demonstrates that principled training design and transparent methodology can yield strong scientific intelligence with substantially reduced data requirements. (i) First, we provide a fully transparent, end-to-end reproducible training pipeline, covering data collection, cleaning, preprocessing, supervised fine-tuning, reinforcement learning, and evaluation, along with detailed optimization recipes. This facilitates systematic extension by the community. (ii) Second, Innovator-VL exhibits remarkable data efficiency, achieving competitive performance on various scientific tasks using fewer than five million curated samples without large-scale pretraining. These results highlight that effective reasoning can be achieved through principled data selection rather than indiscriminate scaling. (iii) Third, Innovator-VL demonstrates strong generalization, achieving competitive performance on general vision, multimodal reasoning, and scientific benchmarks. This indicates that scientific alignment can be integrated into a unified model without compromising general-purpose capabilities. Our practices suggest that efficient, reproducible, and high-performing scientific multimodal models can be built even without large-scale data, providing a practical foundation for future research.
Membership Inference Attack with Partial Features
Aug 08, 2025Abstract:Machine learning models have been shown to be susceptible to membership inference attack, which can be used to determine whether a given sample appears in the training data. Existing membership inference methods commonly assume that the adversary has full access to the features of the target sample. This assumption, however, does not hold in many real-world scenarios where only partial features information is available, thereby limiting the applicability of these methods. In this work, we study an inference scenario where the adversary observes only partial features of each sample and aims to infer whether this observed subset was present in the training set of the target model. We define this problem as Partial Feature Membership Inference (PFMI). To address this problem, we propose MRAD (Memory-guided Reconstruction and Anomaly Detection), a two-stage attack framework. In the first stage, MRAD optimizes the unknown feature values to minimize the loss of the sample. In the second stage, it measures the deviation between the reconstructed sample and the training distribution using anomaly detection. Empirical results demonstrate that MRAD is effective across a range of datasets, and maintains compatibility with various off-the-shelf anomaly detection techniques. For example, on STL-10, our attack achieves an AUC of around 0.6 even with 40% of the missing features.
A Simple Review of EEG Foundation Models: Datasets, Advancements and Future Perspectives
Apr 24, 2025Abstract:Electroencephalogram (EEG) signals play a crucial role in understanding brain activity and diagnosing neurological disorders. This review focuses on the recent development of EEG foundation models(EEG-FMs), which have shown great potential in processing and analyzing EEG data. We discuss various EEG-FMs, including their architectures, pre-training strategies, their pre-training and downstream datasets and other details. The review also highlights the challenges and future directions in this field, aiming to provide a comprehensive overview for researchers and practitioners interested in EEG analysis and related EEG-FMs.
Uni-3DAR: Unified 3D Generation and Understanding via Autoregression on Compressed Spatial Tokens
Mar 21, 2025Abstract:Recent advancements in large language models and their multi-modal extensions have demonstrated the effectiveness of unifying generation and understanding through autoregressive next-token prediction. However, despite the critical role of 3D structural generation and understanding (3D GU) in AI for science, these tasks have largely evolved independently, with autoregressive methods remaining underexplored. To bridge this gap, we introduce Uni-3DAR, a unified framework that seamlessly integrates 3D GU tasks via autoregressive prediction. At its core, Uni-3DAR employs a novel hierarchical tokenization that compresses 3D space using an octree, leveraging the inherent sparsity of 3D structures. It then applies an additional tokenization for fine-grained structural details, capturing key attributes such as atom types and precise spatial coordinates in microscopic 3D structures. We further propose two optimizations to enhance efficiency and effectiveness. The first is a two-level subtree compression strategy, which reduces the octree token sequence by up to 8x. The second is a masked next-token prediction mechanism tailored for dynamically varying token positions, significantly boosting model performance. By combining these strategies, Uni-3DAR successfully unifies diverse 3D GU tasks within a single autoregressive framework. Extensive experiments across multiple microscopic 3D GU tasks, including molecules, proteins, polymers, and crystals, validate its effectiveness and versatility. Notably, Uni-3DAR surpasses previous state-of-the-art diffusion models by a substantial margin, achieving up to 256\% relative improvement while delivering inference speeds up to 21.8x faster. The code is publicly available at https://github.com/dptech-corp/Uni-3DAR.
OPTIMUS: Predicting Multivariate Outcomes in Alzheimer's Disease Using Multi-modal Data amidst Missing Values
Mar 14, 2025Abstract:Alzheimer's disease, a neurodegenerative disorder, is associated with neural, genetic, and proteomic factors while affecting multiple cognitive and behavioral faculties. Traditional AD prediction largely focuses on univariate disease outcomes, such as disease stages and severity. Multimodal data encode broader disease information than a single modality and may, therefore, improve disease prediction; but they often contain missing values. Recent "deeper" machine learning approaches show promise in improving prediction accuracy, yet the biological relevance of these models needs to be further charted. Integrating missing data analysis, predictive modeling, multimodal data analysis, and explainable AI, we propose OPTIMUS, a predictive, modular, and explainable machine learning framework, to unveil the many-to-many predictive pathways between multimodal input data and multivariate disease outcomes amidst missing values. OPTIMUS first applies modality-specific imputation to uncover data from each modality while optimizing overall prediction accuracy. It then maps multimodal biomarkers to multivariate outcomes using machine-learning and extracts biomarkers respectively predictive of each outcome. Finally, OPTIMUS incorporates XAI to explain the identified multimodal biomarkers. Using data from 346 cognitively normal subjects, 608 persons with mild cognitive impairment, and 251 AD patients, OPTIMUS identifies neural and transcriptomic signatures that jointly but differentially predict multivariate outcomes related to executive function, language, memory, and visuospatial function. Our work demonstrates the potential of building a predictive and biologically explainable machine-learning framework to uncover multimodal biomarkers that capture disease profiles across varying cognitive landscapes. The results improve our understanding of the complex many-to-many pathways in AD.
Representation Learning, Large-Scale 3D Molecular Pretraining, Molecular Property
Mar 13, 2025Abstract:Molecular pretrained representations (MPR) has emerged as a powerful approach for addressing the challenge of limited supervised data in applications such as drug discovery and material design. While early MPR methods relied on 1D sequences and 2D graphs, recent advancements have incorporated 3D conformational information to capture rich atomic interactions. However, these prior models treat molecules merely as discrete atom sets, overlooking the space surrounding them. We argue from a physical perspective that only modeling these discrete points is insufficient. We first present a simple yet insightful observation: naively adding randomly sampled virtual points beyond atoms can surprisingly enhance MPR performance. In light of this, we propose a principled framework that incorporates the entire 3D space spanned by molecules. We implement the framework via a novel Transformer-based architecture, dubbed SpaceFormer, with three key components: (1) grid-based space discretization; (2) grid sampling/merging; and (3) efficient 3D positional encoding. Extensive experiments show that SpaceFormer significantly outperforms previous 3D MPR models across various downstream tasks with limited data, validating the benefit of leveraging the additional 3D space beyond atoms in MPR models.
MolParser: End-to-end Visual Recognition of Molecule Structures in the Wild
Nov 17, 2024



Abstract:In recent decades, chemistry publications and patents have increased rapidly. A significant portion of key information is embedded in molecular structure figures, complicating large-scale literature searches and limiting the application of large language models in fields such as biology, chemistry, and pharmaceuticals. The automatic extraction of precise chemical structures is of critical importance. However, the presence of numerous Markush structures in real-world documents, along with variations in molecular image quality, drawing styles, and noise, significantly limits the performance of existing optical chemical structure recognition (OCSR) methods. We present MolParser, a novel end-to-end OCSR method that efficiently and accurately recognizes chemical structures from real-world documents, including difficult Markush structure. We use a extended SMILES encoding rule to annotate our training dataset. Under this rule, we build MolParser-7M, the largest annotated molecular image dataset to our knowledge. While utilizing a large amount of synthetic data, we employed active learning methods to incorporate substantial in-the-wild data, specifically samples cropped from real patents and scientific literature, into the training process. We trained an end-to-end molecular image captioning model, MolParser, using a curriculum learning approach. MolParser significantly outperforms classical and learning-based methods across most scenarios, with potential for broader downstream applications. The dataset is publicly available.
Copiloting Diagnosis of Autism in Real Clinical Scenarios via LLMs
Oct 10, 2024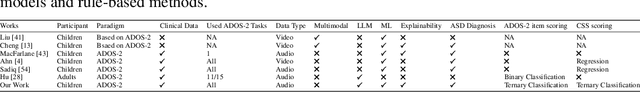
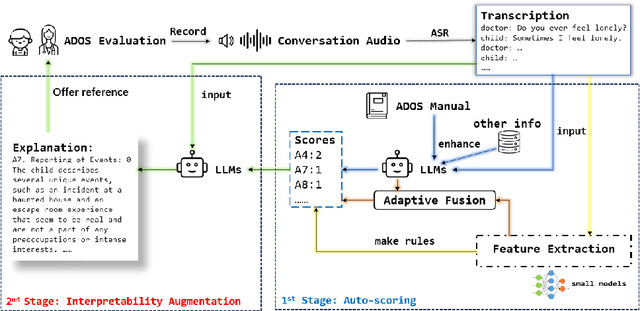
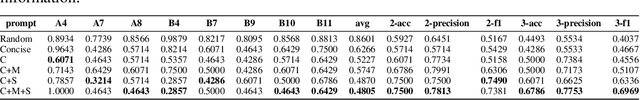

Abstract:Autism spectrum disorder(ASD) is a pervasive developmental disorder that significantly impacts the daily functioning and social participation of individuals. Despite the abundance of research focused on supporting the clinical diagnosis of ASD, there is still a lack of systematic and comprehensive exploration in the field of methods based on Large Language Models (LLMs), particularly regarding the real-world clinical diagnostic scenarios based on Autism Diagnostic Observation Schedule, Second Edition (ADOS-2). Therefore, we have proposed a framework called ADOS-Copilot, which strikes a balance between scoring and explanation and explored the factors that influence the performance of LLMs in this task. The experimental results indicate that our proposed framework is competitive with the diagnostic results of clinicians, with a minimum MAE of 0.4643, binary classification F1-score of 81.79\%, and ternary classification F1-score of 78.37\%. Furthermore, we have systematically elucidated the strengths and limitations of current LLMs in this task from the perspectives of ADOS-2, LLMs' capabilities, language, and model scale aiming to inspire and guide the future application of LLMs in a broader fields of mental health disorders. We hope for more research to be transferred into real clinical practice, opening a window of kindness to the world for eccentric children.
A high-accuracy multi-model mixing retrosynthetic method
Sep 06, 2024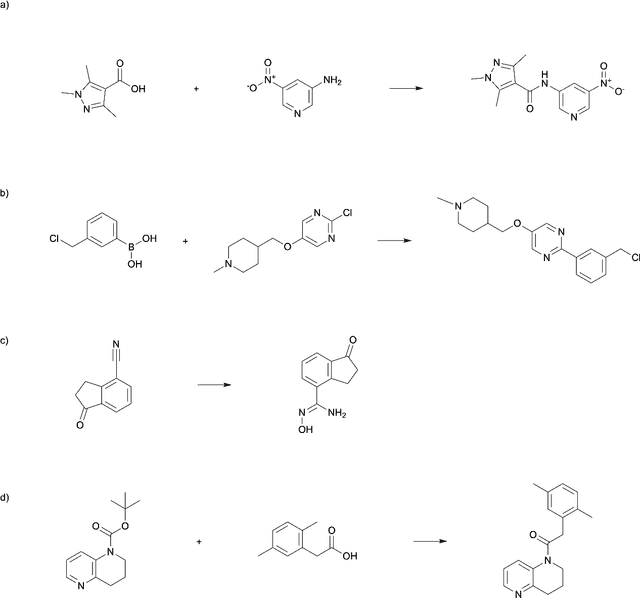
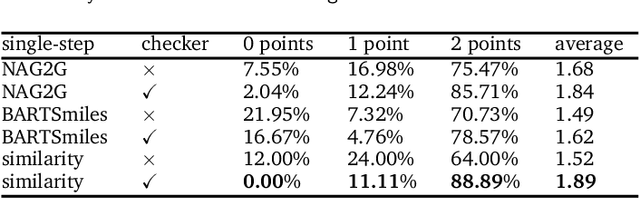
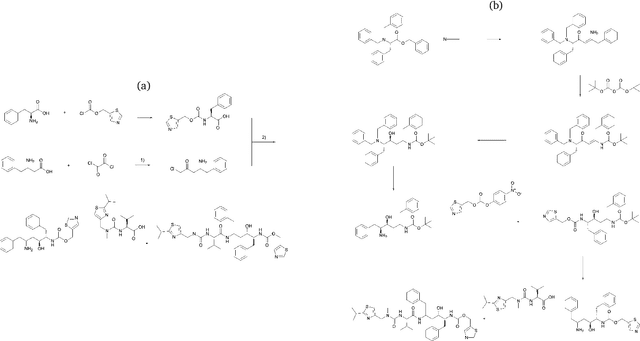
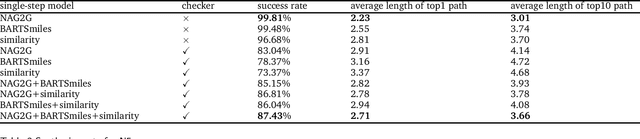
Abstract:The field of computer-aided synthesis planning (CASP) has seen rapid advancements in recent years, achieving significant progress across various algorithmic benchmarks. However, chemists often encounter numerous infeasible reactions when using CASP in practice. This article delves into common errors associated with CASP and introduces a product prediction model aimed at enhancing the accuracy of single-step models. While the product prediction model reduces the number of single-step reactions, it integrates multiple single-step models to maintain the overall reaction count and increase reaction diversity. Based on manual analysis and large-scale testing, the product prediction model, combined with the multi-model ensemble approach, has been proven to offer higher feasibility and greater diversity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge