Oliver Y. Chén
REMEMBER: Retrieval-based Explainable Multimodal Evidence-guided Modeling for Brain Evaluation and Reasoning in Zero- and Few-shot Neurodegenerative Diagnosis
Apr 12, 2025Abstract:Timely and accurate diagnosis of neurodegenerative disorders, such as Alzheimer's disease, is central to disease management. Existing deep learning models require large-scale annotated datasets and often function as "black boxes". Additionally, datasets in clinical practice are frequently small or unlabeled, restricting the full potential of deep learning methods. Here, we introduce REMEMBER -- Retrieval-based Explainable Multimodal Evidence-guided Modeling for Brain Evaluation and Reasoning -- a new machine learning framework that facilitates zero- and few-shot Alzheimer's diagnosis using brain MRI scans through a reference-based reasoning process. Specifically, REMEMBER first trains a contrastively aligned vision-text model using expert-annotated reference data and extends pseudo-text modalities that encode abnormality types, diagnosis labels, and composite clinical descriptions. Then, at inference time, REMEMBER retrieves similar, human-validated cases from a curated dataset and integrates their contextual information through a dedicated evidence encoding module and attention-based inference head. Such an evidence-guided design enables REMEMBER to imitate real-world clinical decision-making process by grounding predictions in retrieved imaging and textual context. Specifically, REMEMBER outputs diagnostic predictions alongside an interpretable report, including reference images and explanations aligned with clinical workflows. Experimental results demonstrate that REMEMBER achieves robust zero- and few-shot performance and offers a powerful and explainable framework to neuroimaging-based diagnosis in the real world, especially under limited data.
Explainable Graph-theoretical Machine Learning: with Application to Alzheimer's Disease Prediction
Mar 20, 2025



Abstract:Alzheimer's disease (AD) affects 50 million people worldwide and is projected to overwhelm 152 million by 2050. AD is characterized by cognitive decline due partly to disruptions in metabolic brain connectivity. Thus, early and accurate detection of metabolic brain network impairments is crucial for AD management. Chief to identifying such impairments is FDG-PET data. Despite advancements, most graph-based studies using FDG-PET data rely on group-level analysis or thresholding. Yet, group-level analysis can veil individual differences and thresholding may overlook weaker but biologically critical brain connections. Additionally, machine learning-based AD prediction largely focuses on univariate outcomes, such as disease status. Here, we introduce explainable graph-theoretical machine learning (XGML), a framework employing kernel density estimation and dynamic time warping to construct individual metabolic brain graphs that capture the distance between pair-wise brain regions and identify subgraphs most predictive of multivariate AD-related outcomes. Using FDG-PET data from the Alzheimer's Disease Neuroimaging Initiative, XGML builds metabolic brain graphs and uncovers subgraphs predictive of eight AD-related cognitive scores in new subjects. XGML shows robust performance, particularly for predicting scores measuring learning, memory, language, praxis, and orientation, such as CDRSB ($r = 0.74$), ADAS11 ($r = 0.73$), and ADAS13 ($r = 0.71$). Moreover, XGML unveils key edges jointly but differentially predictive of several AD-related outcomes; they may serve as potential network biomarkers for assessing overall cognitive decline. Together, we show the promise of graph-theoretical machine learning in biomarker discovery and disease prediction and its potential to improve our understanding of network neural mechanisms underlying AD.
OPTIMUS: Predicting Multivariate Outcomes in Alzheimer's Disease Using Multi-modal Data amidst Missing Values
Mar 14, 2025Abstract:Alzheimer's disease, a neurodegenerative disorder, is associated with neural, genetic, and proteomic factors while affecting multiple cognitive and behavioral faculties. Traditional AD prediction largely focuses on univariate disease outcomes, such as disease stages and severity. Multimodal data encode broader disease information than a single modality and may, therefore, improve disease prediction; but they often contain missing values. Recent "deeper" machine learning approaches show promise in improving prediction accuracy, yet the biological relevance of these models needs to be further charted. Integrating missing data analysis, predictive modeling, multimodal data analysis, and explainable AI, we propose OPTIMUS, a predictive, modular, and explainable machine learning framework, to unveil the many-to-many predictive pathways between multimodal input data and multivariate disease outcomes amidst missing values. OPTIMUS first applies modality-specific imputation to uncover data from each modality while optimizing overall prediction accuracy. It then maps multimodal biomarkers to multivariate outcomes using machine-learning and extracts biomarkers respectively predictive of each outcome. Finally, OPTIMUS incorporates XAI to explain the identified multimodal biomarkers. Using data from 346 cognitively normal subjects, 608 persons with mild cognitive impairment, and 251 AD patients, OPTIMUS identifies neural and transcriptomic signatures that jointly but differentially predict multivariate outcomes related to executive function, language, memory, and visuospatial function. Our work demonstrates the potential of building a predictive and biologically explainable machine-learning framework to uncover multimodal biomarkers that capture disease profiles across varying cognitive landscapes. The results improve our understanding of the complex many-to-many pathways in AD.
VisTA: Vision-Text Alignment Model with Contrastive Learning using Multimodal Data for Evidence-Driven, Reliable, and Explainable Alzheimer's Disease Diagnosis
Feb 03, 2025Abstract:Objective: Assessing Alzheimer's disease (AD) using high-dimensional radiology images is clinically important but challenging. Although Artificial Intelligence (AI) has advanced AD diagnosis, it remains unclear how to design AI models embracing predictability and explainability. Here, we propose VisTA, a multimodal language-vision model assisted by contrastive learning, to optimize disease prediction and evidence-based, interpretable explanations for clinical decision-making. Methods: We developed VisTA (Vision-Text Alignment Model) for AD diagnosis. Architecturally, we built VisTA from BiomedCLIP and fine-tuned it using contrastive learning to align images with verified abnormalities and their descriptions. To train VisTA, we used a constructed reference dataset containing images, abnormality types, and descriptions verified by medical experts. VisTA produces four outputs: predicted abnormality type, similarity to reference cases, evidence-driven explanation, and final AD diagnoses. To illustrate VisTA's efficacy, we reported accuracy metrics for abnormality retrieval and dementia prediction. To demonstrate VisTA's explainability, we compared its explanations with human experts' explanations. Results: Compared to 15 million images used for baseline pretraining, VisTA only used 170 samples for fine-tuning and obtained significant improvement in abnormality retrieval and dementia prediction. For abnormality retrieval, VisTA reached 74% accuracy and an AUC of 0.87 (26% and 0.74, respectively, from baseline models). For dementia prediction, VisTA achieved 88% accuracy and an AUC of 0.82 (30% and 0.57, respectively, from baseline models). The generated explanations agreed strongly with human experts' and provided insights into the diagnostic process. Taken together, VisTA optimize prediction, clinical reasoning, and explanation.
Enhancing Uncertain Demand Prediction in Hospitals Using Simple and Advanced Machine Learning
Apr 29, 2024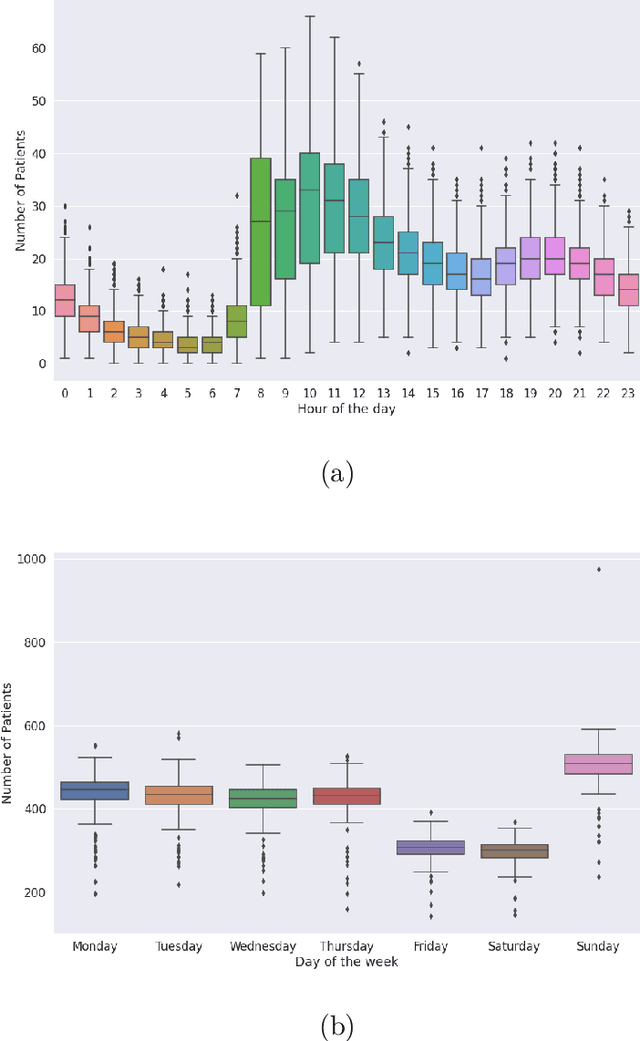
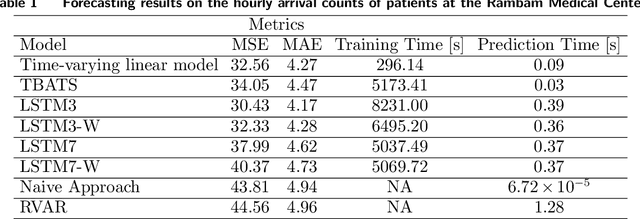
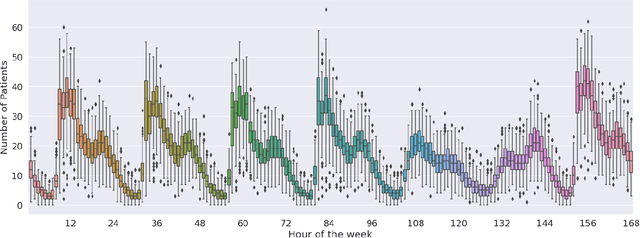
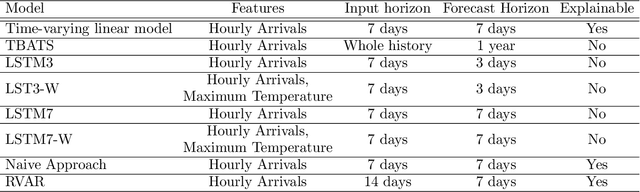
Abstract:Early and timely prediction of patient care demand not only affects effective resource allocation but also influences clinical decision-making as well as patient experience. Accurately predicting patient care demand, however, is a ubiquitous challenge for hospitals across the world due, in part, to the demand's time-varying temporal variability, and, in part, to the difficulty in modelling trends in advance. To address this issue, here, we develop two methods, a relatively simple time-vary linear model, and a more advanced neural network model. The former forecasts patient arrivals hourly over a week based on factors such as day of the week and previous 7-day arrival patterns. The latter leverages a long short-term memory (LSTM) model, capturing non-linear relationships between past data and a three-day forecasting window. We evaluate the predictive capabilities of the two proposed approaches compared to two na\"ive approaches - a reduced-rank vector autoregressive (VAR) model and the TBATS model. Using patient care demand data from Rambam Medical Center in Israel, our results show that both proposed models effectively capture hourly variations of patient demand. Additionally, the linear model is more explainable thanks to its simple architecture, whereas, by accurately modelling weekly seasonal trends, the LSTM model delivers lower prediction errors. Taken together, our explorations suggest the utility of machine learning in predicting time-varying patient care demand; additionally, it is possible to predict patient care demand with good accuracy (around 4 patients) three days or a week in advance using machine learning.
L-SeqSleepNet: Whole-cycle Long Sequence Modelling for Automatic Sleep Staging
Jan 25, 2023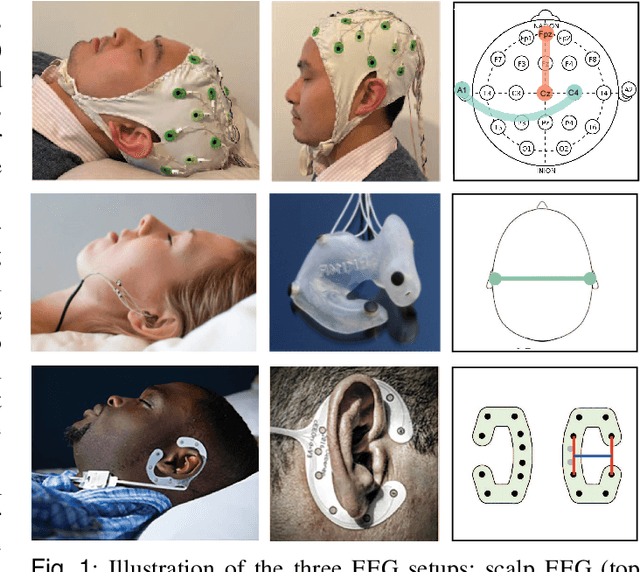
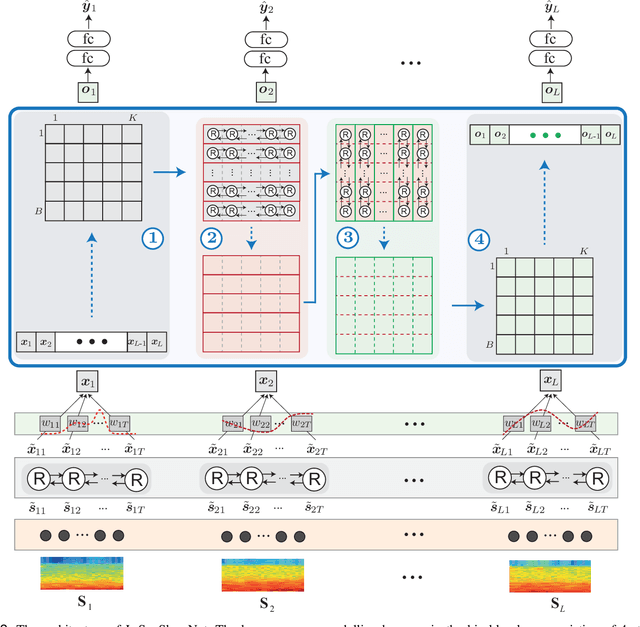
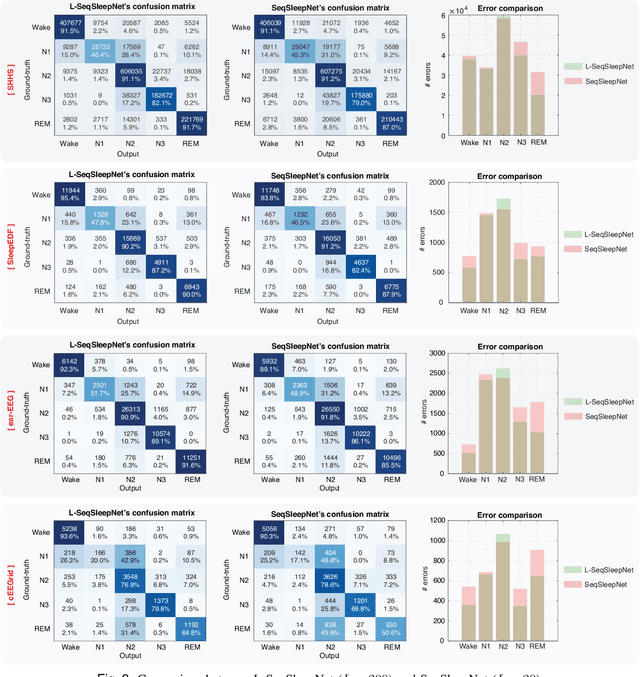
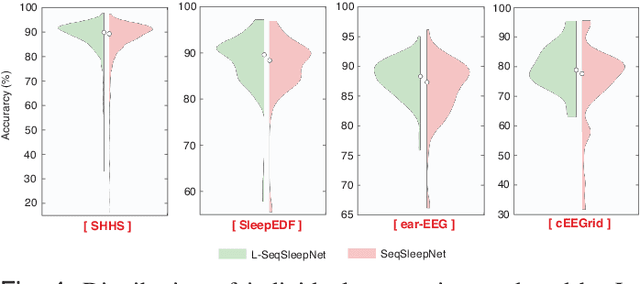
Abstract:Human sleep is cyclical with a period of approximately 90 minutes, implying long temporal dependency in the sleep data. Yet, exploring this long-term dependency when developing sleep staging models has remained untouched. In this work, we show that while encoding the logic of a whole sleep cycle is crucial to improve sleep staging performance, the sequential modelling approach in existing state-of-the-art deep learning models are inefficient for that purpose. We thus introduce a method for efficient long sequence modelling and propose a new deep learning model, L-SeqSleepNet, which takes into account whole-cycle sleep information for sleep staging. Evaluating L-SeqSleepNet on four distinct databases of various sizes, we demonstrate state-of-the-art performance obtained by the model over three different EEG setups, including scalp EEG in conventional Polysomnography (PSG), in-ear EEG, and around-the-ear EEG (cEEGrid), even with a single EEG channel input. Our analyses also show that L-SeqSleepNet is able to alleviate the predominance of N2 sleep (the major class in terms of classification) to bring down errors in other sleep stages. Moreover the network becomes much more robust, meaning that for all subjects where the baseline method had exceptionally poor performance, their performance are improved significantly. Finally, the computation time only grows at a sub-linear rate when the sequence length increases.
Personalized Longitudinal Assessment of Multiple Sclerosis Using Smartphones
Sep 20, 2022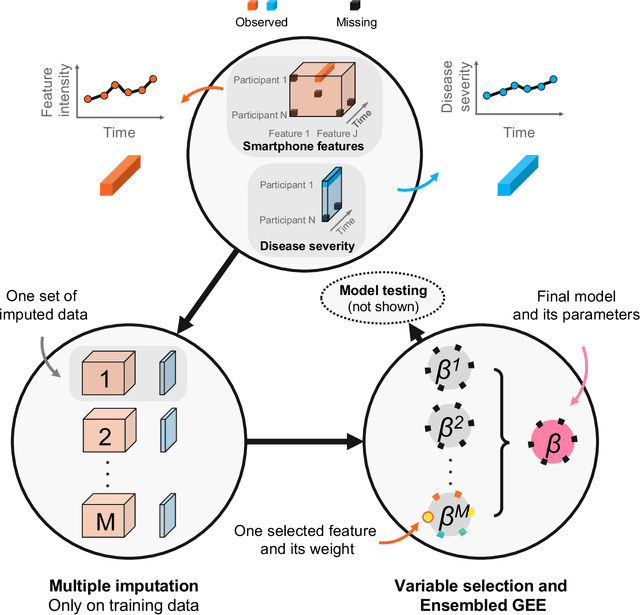
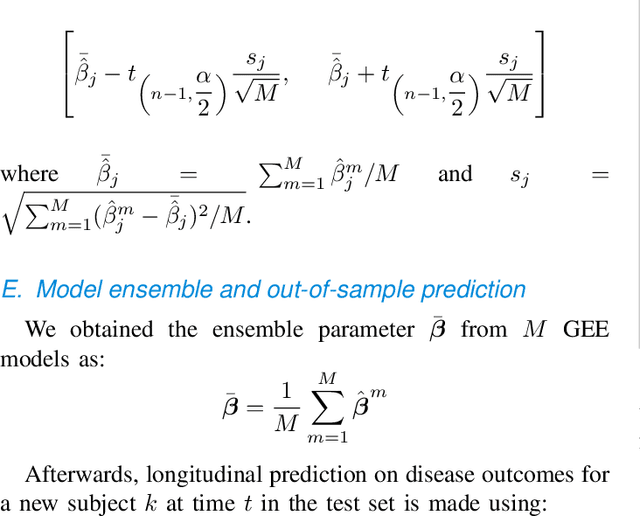

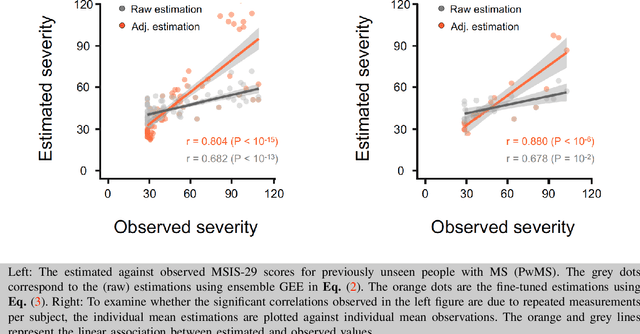
Abstract:Personalized longitudinal disease assessment is central to quickly diagnosing, appropriately managing, and optimally adapting the therapeutic strategy of multiple sclerosis (MS). It is also important for identifying the idiosyncratic subject-specific disease profiles. Here, we design a novel longitudinal model to map individual disease trajectories in an automated way using sensor data that may contain missing values. First, we collect digital measurements related to gait and balance, and upper extremity functions using sensor-based assessments administered on a smartphone. Next, we treat missing data via imputation. We then discover potential markers of MS by employing a generalized estimation equation. Subsequently, parameters learned from multiple training datasets are ensembled to form a simple, unified longitudinal predictive model to forecast MS over time in previously unseen people with MS. To mitigate potential underestimation for individuals with severe disease scores, the final model incorporates additional subject-specific fine-tuning using data from the first day. The results show that the proposed model is promising to achieve personalized longitudinal MS assessment; they also suggest that features related to gait and balance as well as upper extremity function, remotely collected from sensor-based assessments, may be useful digital markers for predicting MS over time.
SleepTransformer: Automatic Sleep Staging with Interpretability and Uncertainty Quantification
May 23, 2021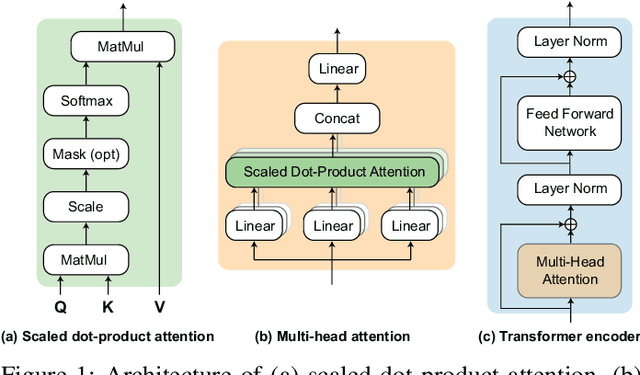
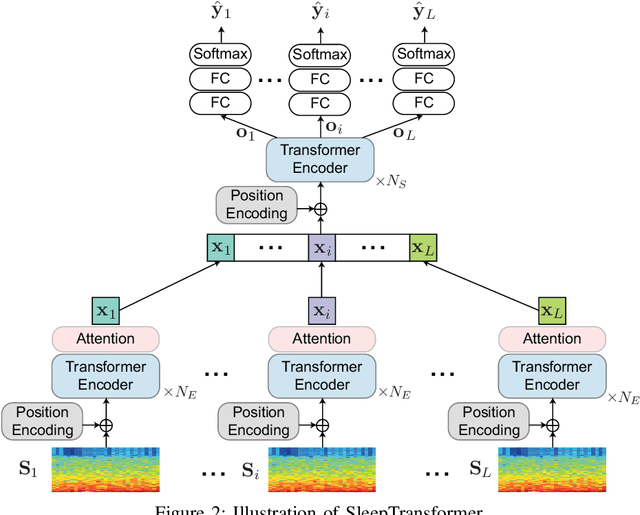

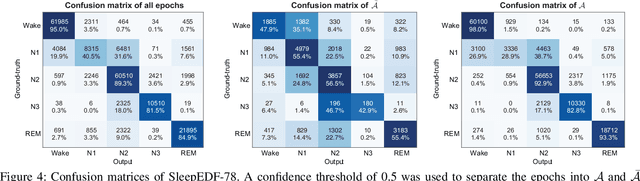
Abstract:Black-box skepticism is one of the main hindrances impeding deep-learning-based automatic sleep scoring from being used in clinical environments. Towards interpretability, this work proposes a sequence-to-sequence sleep-staging model, namely SleepTransformer. It is based on the transformer backbone whose self-attention scores offer interpretability of the model's decisions at both the epoch and sequence level. At the epoch level, the attention scores can be encoded as a heat map to highlight sleep-relevant features captured from the input EEG signal. At the sequence level, the attention scores are visualized as the influence of different neighboring epochs in an input sequence (i.e. the context) to recognition of a target epoch, mimicking the way manual scoring is done by human experts. We further propose a simple yet efficient method to quantify uncertainty in the model's decisions. The method, which is based on entropy, can serve as a metric for deferring low-confidence epochs to a human expert for further inspection. Additionally, we demonstrate that the proposed SleepTransformer outperforms existing methods at a lower computational cost and achieves state-of-the-art performance on two experimental databases of different sizes.
Multi-view Audio and Music Classification
Mar 03, 2021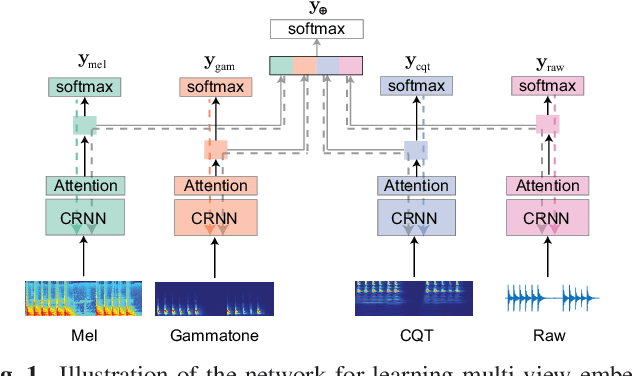

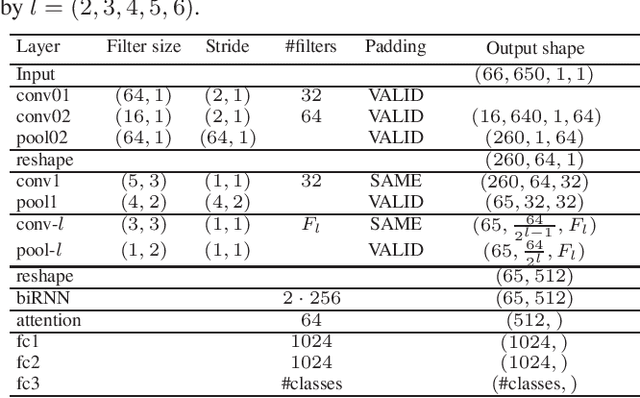

Abstract:We propose in this work a multi-view learning approach for audio and music classification. Considering four typical low-level representations (i.e. different views) commonly used for audio and music recognition tasks, the proposed multi-view network consists of four subnetworks, each handling one input types. The learned embedding in the subnetworks are then concatenated to form the multi-view embedding for classification similar to a simple concatenation network. However, apart from the joint classification branch, the network also maintains four classification branches on the single-view embedding of the subnetworks. A novel method is then proposed to keep track of the learning behavior on the classification branches and adapt their weights to proportionally blend their gradients for network training. The weights are adapted in such a way that learning on a branch that is generalizing well will be encouraged whereas learning on a branch that is overfitting will be slowed down. Experiments on three different audio and music classification tasks show that the proposed multi-view network not only outperforms the single-view baselines but also is superior to the multi-view baselines based on concatenation and late fusion.
Self-Attention Generative Adversarial Network for Speech Enhancement
Oct 18, 2020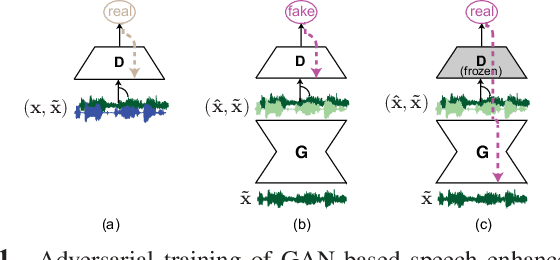
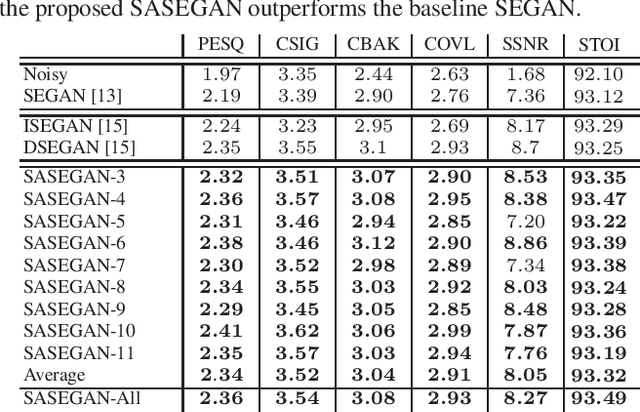
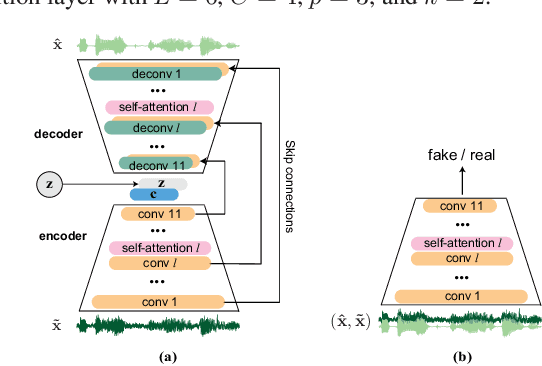
Abstract:Existing generative adversarial networks (GANs) for speech enhancement solely rely on the convolution operation, which may obscure temporal dependencies across the sequence input. To remedy this issue, we propose a self-attention layer adapted from non-local attention, coupled with the convolutional and deconvolutional layers of a speech enhancement GAN (SEGAN) using raw signal input. Further, we empirically study the effect of placing the self-attention layer at the (de)convolutional layers with varying layer indices as well as at all of them when memory allows. Our experiments show that introducing self-attention to SEGAN leads to consistent improvement across the objective evaluation metrics of enhancement performance. Furthermore, applying at different (de)convolutional layers does not significantly alter performance, suggesting that it can be conveniently applied at the highest-level (de)convolutional layer with the smallest memory overhead.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge