Shuwen Yang
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
Uni-Parser Technical Report
Dec 17, 2025Abstract:This technical report introduces Uni-Parser, an industrial-grade document parsing engine tailored for scientific literature and patents, delivering high throughput, robust accuracy, and cost efficiency. Unlike pipeline-based document parsing methods, Uni-Parser employs a modular, loosely coupled multi-expert architecture that preserves fine-grained cross-modal alignments across text, equations, tables, figures, and chemical structures, while remaining easily extensible to emerging modalities. The system incorporates adaptive GPU load balancing, distributed inference, dynamic module orchestration, and configurable modes that support either holistic or modality-specific parsing. Optimized for large-scale cloud deployment, Uni-Parser achieves a processing rate of up to 20 PDF pages per second on 8 x NVIDIA RTX 4090D GPUs, enabling cost-efficient inference across billions of pages. This level of scalability facilitates a broad spectrum of downstream applications, ranging from literature retrieval and summarization to the extraction of chemical structures, reaction schemes, and bioactivity data, as well as the curation of large-scale corpora for training next-generation large language models and AI4Science models.
MolParser: End-to-end Visual Recognition of Molecule Structures in the Wild
Nov 17, 2024



Abstract:In recent decades, chemistry publications and patents have increased rapidly. A significant portion of key information is embedded in molecular structure figures, complicating large-scale literature searches and limiting the application of large language models in fields such as biology, chemistry, and pharmaceuticals. The automatic extraction of precise chemical structures is of critical importance. However, the presence of numerous Markush structures in real-world documents, along with variations in molecular image quality, drawing styles, and noise, significantly limits the performance of existing optical chemical structure recognition (OCSR) methods. We present MolParser, a novel end-to-end OCSR method that efficiently and accurately recognizes chemical structures from real-world documents, including difficult Markush structure. We use a extended SMILES encoding rule to annotate our training dataset. Under this rule, we build MolParser-7M, the largest annotated molecular image dataset to our knowledge. While utilizing a large amount of synthetic data, we employed active learning methods to incorporate substantial in-the-wild data, specifically samples cropped from real patents and scientific literature, into the training process. We trained an end-to-end molecular image captioning model, MolParser, using a curriculum learning approach. MolParser significantly outperforms classical and learning-based methods across most scenarios, with potential for broader downstream applications. The dataset is publicly available.
SciAssess: Benchmarking LLM Proficiency in Scientific Literature Analysis
Mar 15, 2024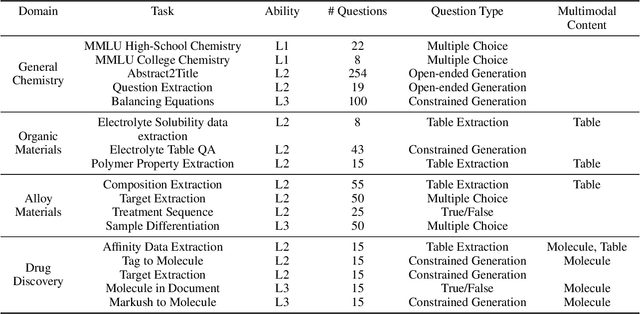
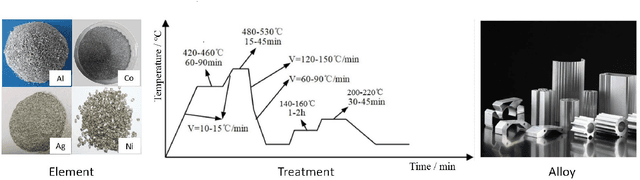
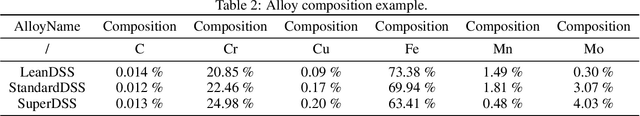
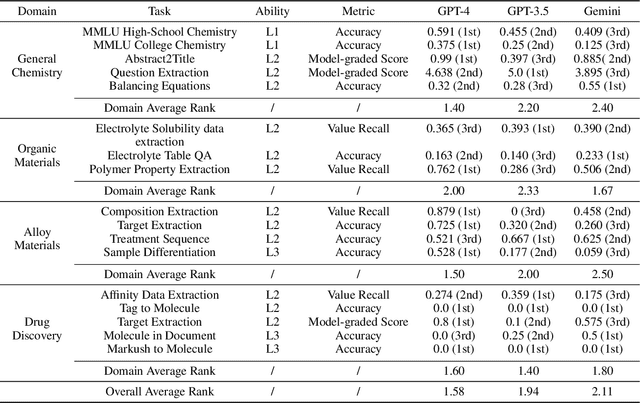
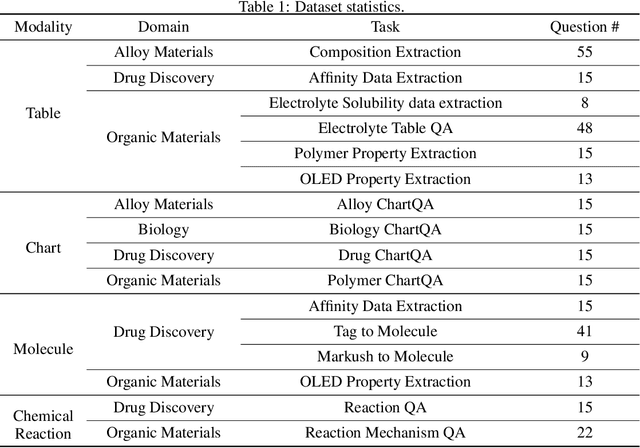
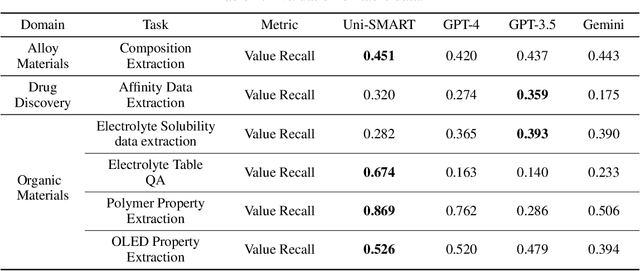
Abstract:Recent breakthroughs in Large Language Models (LLMs) have revolutionized natural language understanding and generation, igniting a surge of interest in leveraging these technologies in the field of scientific literature analysis. Existing benchmarks, however, inadequately evaluate the proficiency of LLMs in scientific literature analysis, especially in scenarios involving complex comprehension and multimodal data. In response, we introduced SciAssess, a benchmark tailored for the in-depth analysis of scientific literature, crafted to provide a thorough assessment of LLMs' efficacy. SciAssess focuses on evaluating LLMs' abilities in memorization, comprehension, and analysis within the context of scientific literature analysis. It includes representative tasks from diverse scientific fields, such as general chemistry, organic materials, and alloy materials. And rigorous quality control measures ensure its reliability in terms of correctness, anonymization, and copyright compliance. SciAssess evaluates leading LLMs, including GPT-4, GPT-3.5, and Gemini, identifying their strengths and aspects for improvement and supporting the ongoing development of LLM applications in scientific literature analysis. SciAssess and its resources are made available at https://sci-assess.github.io, offering a valuable tool for advancing LLM capabilities in scientific literature analysis.
Uni-SMART: Universal Science Multimodal Analysis and Research Transformer
Mar 15, 2024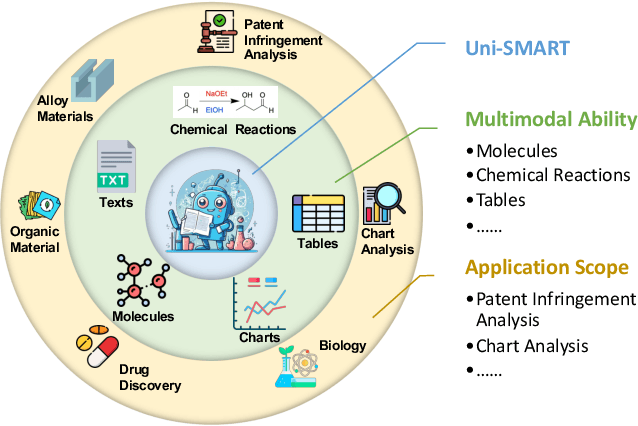
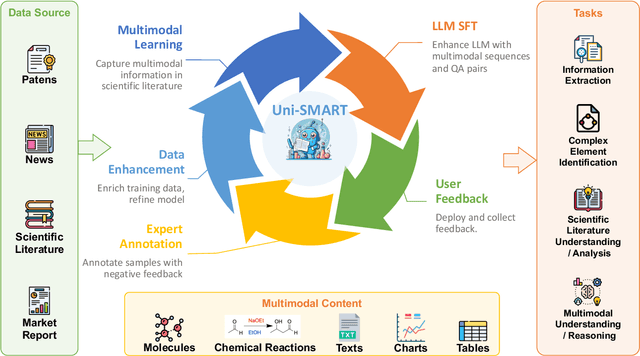
Abstract:In scientific research and its application, scientific literature analysis is crucial as it allows researchers to build on the work of others. However, the fast growth of scientific knowledge has led to a massive increase in scholarly articles, making in-depth literature analysis increasingly challenging and time-consuming. The emergence of Large Language Models (LLMs) has offered a new way to address this challenge. Known for their strong abilities in summarizing texts, LLMs are seen as a potential tool to improve the analysis of scientific literature. However, existing LLMs have their own limits. Scientific literature often includes a wide range of multimodal elements, such as molecular structure, tables, and charts, which are hard for text-focused LLMs to understand and analyze. This issue points to the urgent need for new solutions that can fully understand and analyze multimodal content in scientific literature. To answer this demand, we present Uni-SMART (Universal Science Multimodal Analysis and Research Transformer), an innovative model designed for in-depth understanding of multimodal scientific literature. Through rigorous quantitative evaluation across several domains, Uni-SMART demonstrates superior performance over leading text-focused LLMs. Furthermore, our exploration extends to practical applications, including patent infringement detection and nuanced analysis of charts. These applications not only highlight Uni-SMART's adaptability but also its potential to revolutionize how we interact with scientific literature.
DCQA: Document-Level Chart Question Answering towards Complex Reasoning and Common-Sense Understanding
Oct 29, 2023
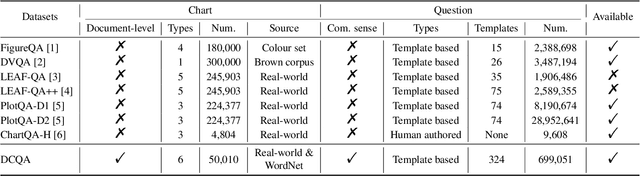
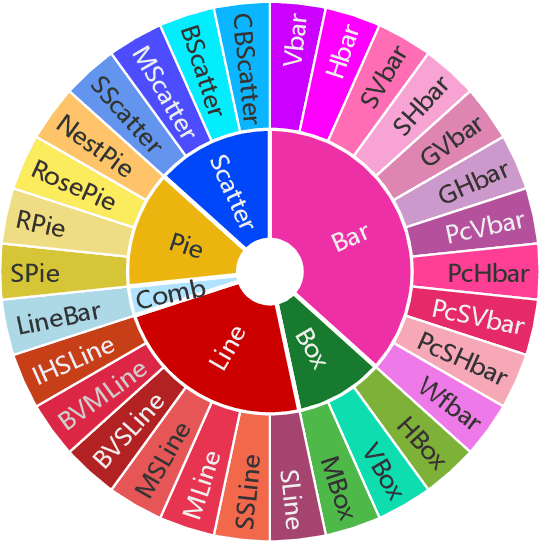
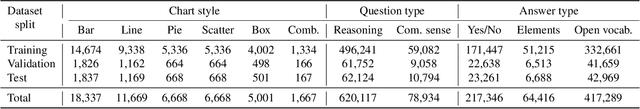
Abstract:Visually-situated languages such as charts and plots are omnipresent in real-world documents. These graphical depictions are human-readable and are often analyzed in visually-rich documents to address a variety of questions that necessitate complex reasoning and common-sense responses. Despite the growing number of datasets that aim to answer questions over charts, most only address this task in isolation, without considering the broader context of document-level question answering. Moreover, such datasets lack adequate common-sense reasoning information in their questions. In this work, we introduce a novel task named document-level chart question answering (DCQA). The goal of this task is to conduct document-level question answering, extracting charts or plots in the document via document layout analysis (DLA) first and subsequently performing chart question answering (CQA). The newly developed benchmark dataset comprises 50,010 synthetic documents integrating charts in a wide range of styles (6 styles in contrast to 3 for PlotQA and ChartQA) and includes 699,051 questions that demand a high degree of reasoning ability and common-sense understanding. Besides, we present the development of a potent question-answer generation engine that employs table data, a rich color set, and basic question templates to produce a vast array of reasoning question-answer pairs automatically. Based on DCQA, we devise an OCR-free transformer for document-level chart-oriented understanding, capable of DLA and answering complex reasoning and common-sense questions over charts in an OCR-free manner. Our DCQA dataset is expected to foster research on understanding visualizations in documents, especially for scenarios that require complex reasoning for charts in the visually-rich document. We implement and evaluate a set of baselines, and our proposed method achieves comparable results.
Progressive Evidence Refinement for Open-domain Multimodal Retrieval Question Answering
Oct 15, 2023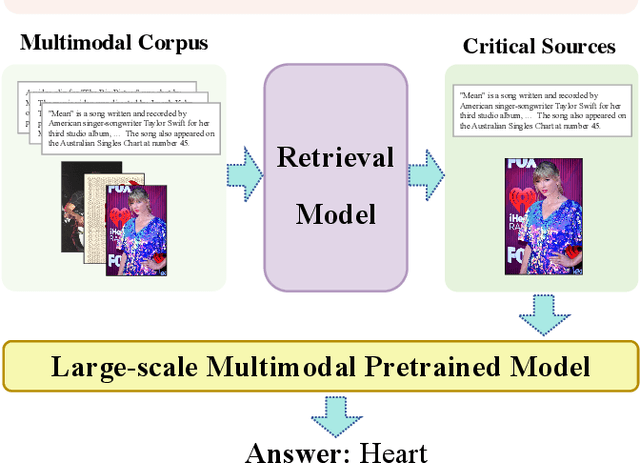
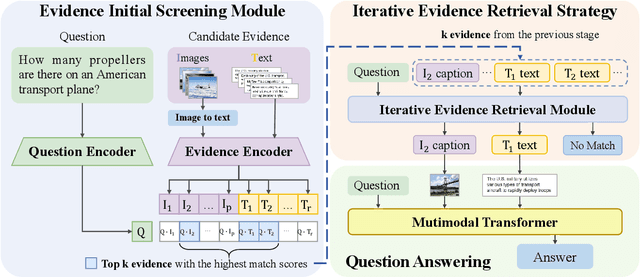
Abstract:Pre-trained multimodal models have achieved significant success in retrieval-based question answering. However, current multimodal retrieval question-answering models face two main challenges. Firstly, utilizing compressed evidence features as input to the model results in the loss of fine-grained information within the evidence. Secondly, a gap exists between the feature extraction of evidence and the question, which hinders the model from effectively extracting critical features from the evidence based on the given question. We propose a two-stage framework for evidence retrieval and question-answering to alleviate these issues. First and foremost, we propose a progressive evidence refinement strategy for selecting crucial evidence. This strategy employs an iterative evidence retrieval approach to uncover the logical sequence among the evidence pieces. It incorporates two rounds of filtering to optimize the solution space, thus further ensuring temporal efficiency. Subsequently, we introduce a semi-supervised contrastive learning training strategy based on negative samples to expand the scope of the question domain, allowing for a more thorough exploration of latent knowledge within known samples. Finally, in order to mitigate the loss of fine-grained information, we devise a multi-turn retrieval and question-answering strategy to handle multimodal inputs. This strategy involves incorporating multimodal evidence directly into the model as part of the historical dialogue and question. Meanwhile, we leverage a cross-modal attention mechanism to capture the underlying connections between the evidence and the question, and the answer is generated through a decoding generation approach. We validate the model's effectiveness through extensive experiments, achieving outstanding performance on WebQA and MultimodelQA benchmark tests.
Deep Molecular Representation Learning via Fusing Physical and Chemical Information
Nov 28, 2021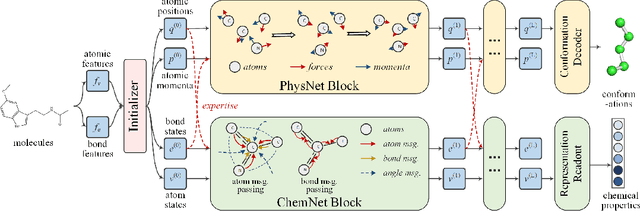
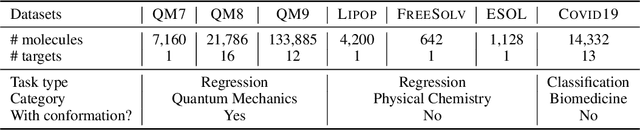
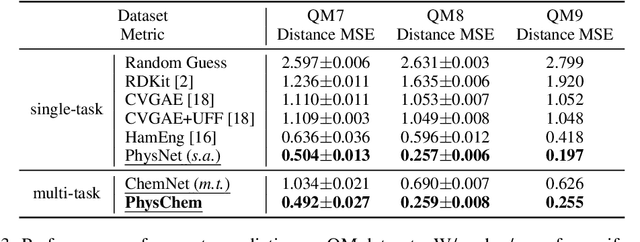
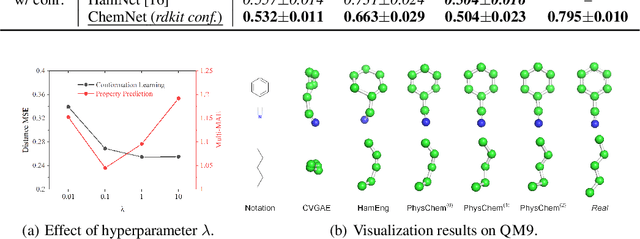
Abstract:Molecular representation learning is the first yet vital step in combining deep learning and molecular science. To push the boundaries of molecular representation learning, we present PhysChem, a novel neural architecture that learns molecular representations via fusing physical and chemical information of molecules. PhysChem is composed of a physicist network (PhysNet) and a chemist network (ChemNet). PhysNet is a neural physical engine that learns molecular conformations through simulating molecular dynamics with parameterized forces; ChemNet implements geometry-aware deep message-passing to learn chemical / biomedical properties of molecules. Two networks specialize in their own tasks and cooperate by providing expertise to each other. By fusing physical and chemical information, PhysChem achieved state-of-the-art performances on MoleculeNet, a standard molecular machine learning benchmark. The effectiveness of PhysChem was further corroborated on cutting-edge datasets of SARS-CoV-2.
HamNet: Conformation-Guided Molecular Representation with Hamiltonian Neural Networks
May 08, 2021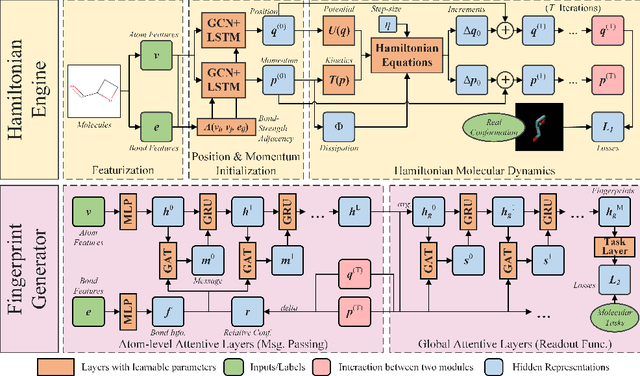
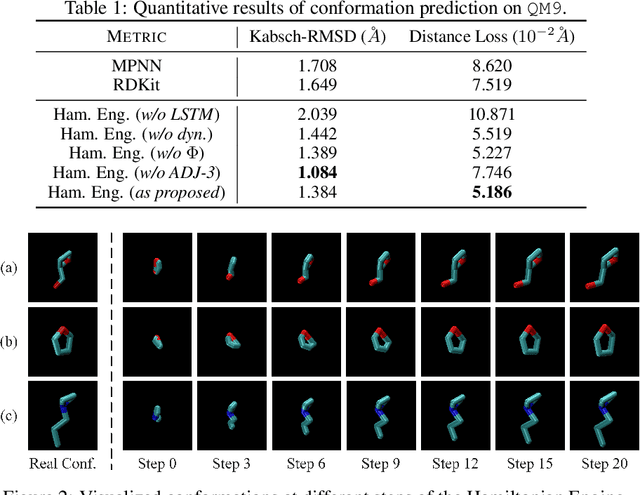
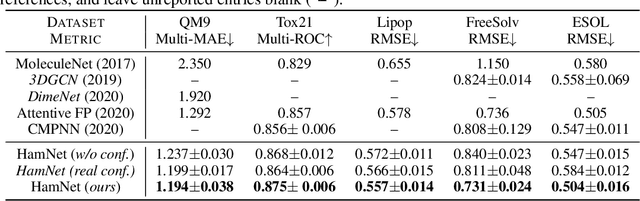
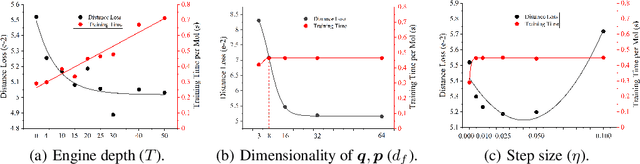
Abstract:Well-designed molecular representations (fingerprints) are vital to combine medical chemistry and deep learning. Whereas incorporating 3D geometry of molecules (i.e. conformations) in their representations seems beneficial, current 3D algorithms are still in infancy. In this paper, we propose a novel molecular representation algorithm which preserves 3D conformations of molecules with a Molecular Hamiltonian Network (HamNet). In HamNet, implicit positions and momentums of atoms in a molecule interact in the Hamiltonian Engine following the discretized Hamiltonian equations. These implicit coordinations are supervised with real conformations with translation- & rotation-invariant losses, and further used as inputs to the Fingerprint Generator, a message-passing neural network. Experiments show that the Hamiltonian Engine can well preserve molecular conformations, and that the fingerprints generated by HamNet achieve state-of-the-art performances on MoleculeNet, a standard molecular machine learning benchmark.
DANE: Domain Adaptive Network Embedding
Jun 03, 2019



Abstract:Recent works reveal that network embedding techniques enable many machine learning models to handle diverse downstream tasks on graph structured data. However, as previous methods usually focus on learning embeddings for a single network, they can not learn representations transferable on multiple networks. Hence, it is important to design a network embedding algorithm that supports downstream model transferring on different networks, known as domain adaptation. In this paper, we propose a novel Domain Adaptive Network Embedding framework, which applies graph convolutional network to learn transferable embeddings. In DANE, nodes from multiple networks are encoded to vectors via a shared set of learnable parameters so that the vectors share an aligned embedding space. The distribution of embeddings on different networks are further aligned by adversarial learning regularization. In addition, DANE's advantage in learning transferable network embedding can be guaranteed theoretically. Extensive experiments reflect that the proposed framework outperforms other state-of-the-art network embedding baselines in cross-network domain adaptation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge